Back to Journals » Infection and Drug Resistance » Volume 11
Molecular detection of vanA and vanB genes among vancomycin-resistant enterococci in ICU-hospitalized patients in Ahvaz in southwest of Iran
Authors Moosavian M , Ghadri H, Samli Z
Received 2 July 2018
Accepted for publication 6 September 2018
Published 15 November 2018 Volume 2018:11 Pages 2269—2275
DOI https://doi.org/10.2147/IDR.S177886
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Mojtaba Moosavian,1,2 Hosein Ghadri,2 Zahra Samli3
1Infectious and Tropical Diseases Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 2Department of Microbiology, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 3School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
Objective: Nosocomial infections due to vancomycin-resistant enterococci (VRE) are known as a source of spreading these bacteria. The aim of this prospective study was molecular detection of vanA and vanB genes among VRE isolated from patients admitted to intensive care units (ICUs) in Ahvaz in southwest of Iran.
Materials and methods: Overall, 243 non-duplicate rectal swab specimens were collected from ICU-hospitalized patients in teaching hospitals affiliated to Ahvaz Jundishapur University of Medical Sciences, Iran. The specimens were inoculated on suitable culture media, and isolates were identified by standard biochemical tests. The susceptibility and resistance of enterococci to 10 antibiotics were determined based on the Clinical and Laboratory Standards Institute guidelines. Resistance to vancomycin was phenotypically detected by vancomycin screening test, and the vanA and vanB genes in vancomycin-resistant isolates were amplified by multiplex PCR method.
Results: Of 175 specimens containing enterococci, 129 (73.7%) isolates were detected as Enterococcus faecium and Enterococcus faecalis and 46 (26.3%) isolates as Enterococcus spp. The results of susceptibility test showed high rates of resistance to tetracycline, erythromycin, ciprofloxacin, and ampicillin. Moreover, based on this test, out of 129 Enterococcus isolates, 56 (43.4%) were resistant to vancomycin and teicoplanin. Also, among 59 vancomycin-resistant or semi-susceptible isolates, vanA gene was detected in 54 (91.5%) isolates, while none of the isolates had vanB gene.
Conclusion: According to the results of this study, to prevent the spread of vancomycin-resistant Enterococcus strains, especially in nosocomial infections, the susceptibility of isolates should be determined before vancomycin prescription.
Keywords: Enterococcus faecalis, Enterococcus faecium, multiplex PCR, vancomycin
Introduction
Enterococci are gram-positive bacteria which are the natural flora of the human intestinal tract.1 Although enterococci are commensal members of the intestinal tract, these organisms can cause nosocomial and community-acquired infections. Urinary tract infections are the most common infections caused by enterococci. These organisms could also be the causative agents of cholecystitis, peritonitis, bacteremia, endocarditis, meningitis, and wound infections.2
These organisms have been reported as the second and third etiologic agents of urinary tract infections and nosocomial bacteremia, respectively.3 Based on a recent report, enterococcal infection occurs in 14.3% of patients admitted to intensive care units (ICUs) during a 24-month period.4
The most common species isolated from clinical samples are Enterococcus faecalis and Enterococcus faecium.5 One of the main reasons that enterococci can survive in hospitals is their inherent resistance to some antibiotics used in hospitals and their ability in acquiring resistance to these common antibiotics.
This resistance has probably been acquired by both mutation and/or receiving resistant genes. In the last decade, the acquisition of resistance to aminoglycosides and glycopeptide antibiotics, especially vancomycin and teicoplanin used in the treatment of enterococci infections, has been common. However, there have been no significant reports on resistance to these antibiotics.6 Six genotypes of resistant enterococci to glycopeptide, including vanA, B, C, D, E, and G have been identified in recent years, the most important of which are vanA and vanB.
VanA and vanB genes are the most common phenotypes observed in hospital isolates. Enterococci containing vanA gene are highly resistant to vancomycin and teicoplanin antibiotics, while enterococci containing vanB gene show high resistance to vancomycin and susceptibility to teicoplanin.2 The hospital prevalence of vancomycin-resistant enterococci (VRE) has been reported in association with the transfer of resistance genes horizontally among the epidemic clones of these bacteria.7
Failure of antibiotic treatments against VRE is a serious and growing problem, resulting in increased mortality and hospital costs.8 For example, the cost of treatment of infections caused by VRE is estimated ten times more than the treatment of enterococci without this resistance.9 A multitude of studies have shown that patients with bacteremia caused by VRE have a higher mortality rate compared with patients with enterococci susceptible to these antibiotics. Patients infected or colonized with VRE are a source for spread of these bacteria, and the presence of VRE in the digestive system of patients is associated with clinically serious infections such as gastrointestinal tract infections.10
Hospitalized patients colonized with VRE are a reservoir for the spread of these bacteria.11 Among the problems caused by VRE are medical restrictions for treating these infections and the ability of these bacteria in the transfer of vancomycin-resistant genes to other gram-positive pathogens. If vancomycin-resistant genes be transferred to the methicillin-resistant Staphylococcus aureus, this pathogen is converted to vancomycin-resistant S. aureus that does not respond to conventional antibiotics.12
Infections caused by VRE in Iran, like other countries, have increased and are followed by mortality, especially in people with weak immune systems.13 This study aimed to evaluate the rate of intestinal colonization of VRE carrying vanA and vanB genes in patients admitted to ICUs in Ahvaz teaching hospitals, Iran.
Materials and methods
In this cross-sectional study, during a period of 12 months, rectal swab specimens were collected from ICU-hospitalized patients at Emam Khomeini and Golestan teaching hospitals affiliated to Ahvaz Jundishapur University of Medical Sciences (AJUMS) in southwest of Iran. These two hospitals are the main hospitals of this city. Ahvaz is one of the major metropolises in Iran, which is considered to be the seventh most populated city in Iran.
Emam Khomeini and Golestan hospitals have 690 and 615 active beds, respectively. On a monthly basis, 20,000 patients are admitted to general clinics and 4,000 patients are hospitalized in these two hospitals.
The active units in these two big hospitals are as follow: ENT, Skin, Oral and Maxillo-Facial Surgery, Infertility, Women-Gynecology, Orthopedics, Urology, Nephrology, Cardiovascular and Pulmonary Section, General and Pediatric Surgery, Physiotherapy, Endoscopy, Angiography, NICU, CCU, ICU, Ophthalmology, Dentistry, Radiotherapy, Nuclear Medicine, MRI, Oncology, Neurology, and other units.
The inclusion criteria were at least one of the following conditions: 1) patients who have been hospitalized for more than 5 days; 2) patients who have received long-term antibiotic vancomycin; 3) patients who have undergone invasive procedures such as catheterization and shunt; and 4) patients with a history of surgery in the chest or abdomen.14 The patients who did not sign the written consent forms or were in a coma were excluded from the study.
Swab specimen collection
In this study, 243 non-duplicate specimens (one per patient) were collected. A sterile swab with a size of 3–5 cm was entered into the patient’s rectum and stirred ~360°; then, the rectal swab was put into a tube containing bile esculin broth (Merck Co, Germany) as a transport medium. The swabs were immediately transferred to the microbiology laboratory of School of Medicine, AJUMS.
The specimens were inoculated on culture media such as blood agar, brain heart infusion (BHI) agar, and bile esculin agar/broth (Merck & Co., Inc.). The plates were incubated at 37°C for 18–24 hours, and the isolated colonies were examined by standard biochemical tests.
Standard biochemical tests
After gram staining, all the isolates were identified by standard biochemical tests such as catalase, 6.5% NaCl, pyrrolidonyl arylamidase (PYR) test, carbohydrates fermentation, motility, pigment production, and vancomycin screening test.15 The gram-positive diplococci, which were negative in catalase and motility tests and positive in esculin hydrolysis and PYR tests, were considered as Enterococcus isolates. Also, these isolates were identified as E. faecalis and E. faecium and other enterococci species based on pigment production on blood agar medium and carbohydrates fermentation such as arabinose, mannitol, sorbitol, sorbose, and lactose.
Antibiotic susceptibility test
The susceptibility and resistance patterns of E. faecalis and E. faecium species to 10 antibiotics were evaluated on Muller Hinton agar (Merck) by the standard Kirby-Bauer disk diffusion method based on the Clinical and Laboratory Standards Institute (CLSI) guidelines.16 In this test, each isolate concentration was equated to 0.5 McFarland, as a standard concentration and the antimicrobial disks (Rosco Diagnostica, Taastrup, Denmark) included: vancomycin (30 µg), teicoplanin (30 µg), ampicillin (10 µg), ciprofloxacin (5 µg), erythromycin (15 µg), tetracycline (30 µg), chloramphenicol (30 µg), fosfomycin (200 µg), linezolid (30 µg), and nitrofurantoin (300 µg).
Vancomycin screening test
This test was performed in order to evaluate phenotypical vancomycin-resistant isolates using BHI agar (Merck) containing 6 µg/mL vancomycin (Sigma-Aldrich Co., St Louis, MO, USA) based on CLSI directions.16
The multiplex PCR method
DNA extraction from Enterococcus isolates was carried out by the boiling method,17 and DNA concentration was measured by biophotometer (Eppendorf, Hamburg, Germany). VanA and vanB genes were amplified using the following primers in a thermocycler (Eppendorf):
A1 5′- GGGAAAACGACAATTGC-3 and A2 5-GTACAATGCGGCCGTTA-3′ with a product size of 732 bp for vanA; B1 5′-ATGGGAAGCCGATAGTC-3′ and B2 5′-GATTTCGTTCCTCGA CC-3′ with a product size of 635 bp for vanB.18
The PCR conditions consisted of a pre-denaturation step at 94°C for 5 minutes, followed by 30 cycles of 45 seconds at 94°C, 45 seconds at 59°C, and 45 seconds at 72°C. A final extension step was performed at 72°C for 5 minutes.
Electrophoresis of PCR products
The PCR products were electrophoresed on 1.5% agarose. The voltage used for electrophoresis was 100 volts for 60 minutes.3 The Gel Documentation System (ProteinSimple, San Jose, CA, USA) was used for viewing and imaging the PCR products on the gel. E. faecium ATCC 51559 and E. faecalis ATCC 51299 were used as positive controls for vanA and vanB genes, respectively. Also, E. faecalis ATCC 29212 was used as the negative control for lacking the vancomycin resistance genes.3,13
Statistical analysis
To analyze the data, chi-squared test was run in SPSS version 16 (SPSS Inc., Chicago, IL, USA). P-value <0.05 was considered statistically significant.
Ethical considerations
This study was confirmed by the Ethics Committee of Ahvaz Jundishapur University of Medical Sciences (No. 1392.173).
Results
In this study, 243 non-duplicate specimens collected from the hospitalized patients were examined. The culture results of 23 rectal swab specimens in esculin broth/agar were negative for Enterococcus growth. Although culture of specimens in esculin medium was positive for 220 rectal swab specimens, the standard biochemical tests confirmed Enterococcus growth only in 175 cases, 95 (54.3%) of which belonged to E. faecium (Table 1). The cases that were not confirmed as Enterococcus were omitted from the study.
  | Table 1 Frequency of 175 different species of Enterococcus isolates from hospitalized patients |
Out of the 243 examined patients, 166 (68%) cases were male and 77 (32%) cases were female.
Antimicrobial susceptibility test was performed by the disk diffusion agar method according to the CLSI guidelines for E. faecium (95 isolates) and E. faecalis (34 isolates). Based on the results of antibiogram test, the most resistant isolates to ciprofloxacin (75.8%) and erythromycin (76.8%) were E. faecium and the most resistant isolate to erythromycin and tetracycline was E. faecalis (52.9% and 73.5%, respectively, Table 2).
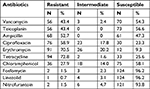  | Table 2 The susceptibility test results of 129 enterococci isolates including 59 Enterococcus faecium and 34 Enterococcus faecalis isolates |
Vancomycin screening test was performed for all Enterococcus isolates that were resistant or semi-susceptible to vancomycin in the disk diffusion agar method. These results showed that out of 59 isolates that were inoculated on BHI agar containing 6 µg/mL vancomycin, 53 isolates grew, which indicated they were fully resistant to vancomycin. We also found that out of 59 resistant or semi-susceptible isolates to vancomycin, 56 and three isolates were identified as E. faecium and E. faecalis, respectively.
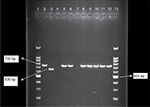
The vanA and vanB genes were amplified in all the resistant or semi-susceptible isolates to vancomycin and teicoplanin by the multiplex PCR method. The results showed that out of 59 isolates, 54 (91.5%) contained vanA gene, and 5 (8.5%) isolates were without vanA and vanB genes (Figure 1). Finally, among 129 patients who were positive for E. faecium and E. faecalis based on the presence of vanA gene in 54 isolates, VRE prevalence was 41.8%. Also, out of 56 E. faecium isolates, 54 contained vanA gene; however, two E. faecium isolates and three E. faecalis isolates did not have vanA gene (P<0.05).
Discussion
Enterococci are opportunist pathogens that could cause nosocomial and community-acquired infections. Among different species, E. faecalis and E. faecium are the most common ones that constitute about 85%–90% of all clinical isolates.19,20 Based on a recent study, 14.3% of patients admitted to ICUs were affected by enterococcal infections.4
In the present study, out of 343 rectal swab specimens, 175 Enterococcus isolates including 95, 34, and 46 isolates of E. faecium, E. faecalis, and Enterococcus spp. were detected, respectively.
In our study, the prevalence rate of E. faecium (54.3%) was more than E. faecalis (19.4%), which is consistent with the study of Sharifi et al in northwest of Iran,21 but it was different from the results of Emaneini et al in Tehran, the capital of Iran and Kafil et al in Tabriz, Iran.22,23 They showed that the prevalence rate of E. faecalis (64.4% and 57%) was more than E. faecium isolates (35.6% and 43%). The reason for these differences in findings could be related to the types of specimens, for example, rectal swabs were used in our study, but other sorts of specimens such as urine, blood, or wound were used in their studies. Since colonization of enterococci in the digestive system is the most common source of spread of VRE,24 we used rectal swabs for sampling in this study. However, the results of some studies in South Korea25 and Switzerland11 were in line with our findings regarding the higher frequency of E. faecium compared to E. faecalis.
Some studies in Iran22 and China26 have indicated that multidrug-resistant enterococci, especially vancomycin-resistant E. faecium isolates, have increased in recent years, which was confirmed in our study. However, among enterococci isolated from different kinds of clinical specimens such as urine, blood, and wound, the prevalence of E. faecalis still surpasses that of E. faecium.27
According to the antibiogram test, all E. faecalis isolates were susceptible to teicoplanin, ampicillin, fosfomycin, linezolid, and nitrofurantoin. A reason for high susceptibility to these antibiotics is probably less prescription of them by physicians. Our results were in agreement with those of Kafil et al in Iran who showed that VRE isolates were susceptible to linezolid, nitrofurantoin, and tigecyclin.23
Our study also revealed high resistance to tetracycline and erythromycin (Table 2), but the resistance rate to these antibiotics for E. faecium was more than that of E. faecium isolates. Also, out of 129 Enterococcus isolates, one isolate was linezolid-resistant and two isolates were resistant to fosfomycin and nitrofurantoin.
The results of our study indicated an increase in vancomycin-resistant E. faecium isolates compared to some studies in Iran, such as the study of Emaneini et al in Tehran.22 The reason for this finding could be related to the studied cases. We studied patients hospitalized in ICUs for more than 5 days, and in some cases, they had received vancomycin for a long time, thus they were at high risk for infection with VRE. Some previous studies in USA28 and Iran29 also showed that the rates of vancomycin-resistant E. faecium isolates were 80% and 100%, respectively. In addition, a recent study in Iran indicated that E. faecium strains were varied among environmental and clinical VRE isolates.30
The phenotypic vancomycin screening test was performed for all enterococci isolates resistant or semi-susceptible to vancomycin in disk-diffusion agar test. Since according to the CLSI criteria, susceptibility of screening test using the BHI medium containing 6 µL/mL vancomycin is more than disk-diffusion agar test, the results of the screening test confirmed that 53 E. faecium isolates were resistant to vancomycin and three semi-susceptible isolates of E. faecalis were identified to be susceptible to vancomycin. However, three E. faecium isolates showed resistance in disk-diffusion agar test, but did not grow in screening test followed by PCR method.
The multiplex PCR assay showed that 54 (91.5%) of Enterococcus isolates contained vanA gene, and none of the isolates contained vanB gene. However, the prevalence of vanA gene among E. faecium and E. faecalis strains was significantly different (P<0.05). The phenotypical resistance of five E. faecium and E. faecalis isolates which had no vanA and vanB genes could be related to different reasons other than genetic agents.
Talebi et al in Iran also showed the presence of vanA gene and the absence of vanB among vancomycin-resistant Enterococcus isolates.8 Similarly, the results of a study in northwest of Iran (2012) showed that 43 VRE strains contained vanA gene, three strains contained vanB gene, and two strains did not have any of the two genes.21 These results were relatively consistent with the present findings, but the slight differences can partially be attributed to the type of specimens and the study period.
However, the findings of Karki et al, in Melbourne, Australia, were not consistent with our results.31 Among 331 rectal specimens, they found 58 (17.5%) VRE containing vanB gene, but none of them had vanA. The reason expressed in their study was that vanB gene in VRE has been endemic in the region. Since unlike vanA gene, vanB is clustered and occupies a large area on the chromosome, the possibility of its transmission among strains is less. Furthermore, vanB gene is mainly associated with epidemics and food contamination, while vanA gene is associated with clinical strains,10 and the fact that patients received vancomycin for a long time justifies the existence of vanA gene in VRE.
Conclusion
Generally, the prevalence of VRE in our hospitals and studied area is increasing. Considering the importance of vancomycin as a bactericidal antibiotic in the treatment of bacterial infections, the outbreak of VRE may create many problems for patients’ health. Through the detection of resistant strains and proper treatment of infected patients, resistance development to vancomycin can be prevented.
Acknowledgments
This research is a part of MSc thesis of Hosein Ghadri which was approved as a research plan (No. 94119) and was financially supported by Deputy of Vice-Chancellor for Research Affairs and Infectious & Tropical Diseases Research Center, AJUMS. The authors thank all of them. The authors express thanks for the patients who participated in this study.
Disclosure
The authors report no conflicts of interest in this work.
References
Harbarth S, Cosgrove S, Carmeli Y. Effects of antibiotics on nosocomial epidemiology of vancomycin-resistant enterococci. Antimicrob Agents Chemother. 2002;46(6):1619–1628. | ||
Nichol KA, Sill M, Laing NM, Johnson JL, Hoban DJ, Zhanel GG. Molecular epidemiology of urinary tract isolates of vancomycin-resistant Enterococcus faecium from North America. Int J Antimicrob Agents. 2006;27(5):392–396. | ||
Yasliani S, Mohabati Mobarez A, Hosseini Doust R, Satari M, Teymornejad O. Linezolid vancomycin resistant Enterococcus isolated from clinical samples in Tehran hospitals. Indian J Med Sci. 2009;63(7):297–302. | ||
Papadimitriou-Olivgeris M, Drougka E, Fligou F, et al. Risk factors for enterococcal infection and colonization by vancomycin-resistant enterococci in critically ill patients. Infection. 2014;42(6):1013–1022. | ||
Panesso D, Reyes J, Rincón S, et al. Molecular epidemiology of vancomycin-resistant Enterococcus faecium: a prospective, multicenter study in South American hospitals. J Clin Microbiol. 2010;48(5):1562–1569. | ||
Shin JW, Yong D, Kim MS, et al. Sudden increase of vancomycin-resistant enterococcal infections in a Korean tertiary care hospital: possible consequences of increased use of oral vancomycin. J Infect Chemother. 2003;9(1):62–67. | ||
Ergani-Ozcan A, Naas T, Baysan BO, et al. Nosocomial outbreak of vancomycin-resistant Enterococcus faecium in a paediatric unit at a Turkish university hospital. J Antimicrob Chemother. 2008;61(5):1033–1039. | ||
Talebi M, Eshraghi SS, Pourshafie MR, Pourmand MR, Eshraghian MR. Characterization of vancomycin resistant faecium. Iranian J Publ Health. 2007;36(4):20–25. | ||
Oliveira AC, Bettcher L. Epidemiological aspects of the occurrence of vancomycin-resistant enterococci. Rev Esc Enferm USP. 2010;44(3):725–731. | ||
Marothi YA, Agnihotri H, Dubey D. Enterococcal resistance--an overview. Indian J Med Microbiol. 2005;23(4):214–219. | ||
Balzereit-Scheuerlein F, Stephan R. Prevalence of colonisation and resistance patterns of vancomycin-resistant enterococci in healthy, non-hospitalised persons in Switzerland. Swiss Med Wkly. 2001;131(19–20):280–282. | ||
Forbes BA, Sahm DF, Weissfeld AS. Bailey & Scott’s Diagnostic Microbiology. 12th ed. St Louis, MI: Mosby, Elsevier; 2007:268–269. | ||
Feizabadi MM, Aliahmadi A, Mobasheri F, Asgharzadeh A, Asadi S, Etemadi G. Phenotypic characteristics and population genetics of Enterococcus faecalis cultured from patients in Tehran during 2000–2001. Can J Microbiol. 2003;49(10):645–649. | ||
D’Azevedo PA, Santiago KA, Furtado GH, Xavier DB, Pignatari AC, Titze-de-Almeida R. Rapid detection of vancomycin-resistant enterococci (VRE) in rectal samples from patients admitted to intensive care units. Braz J Infect Dis. 2009;13(4):289–293. | ||
Procop GW, Church DL, Hall GS, et al; Koneman’s Color Atlas and Textbook of Diagnostic Microbiology. 7th ed. Philadelphia, PA: Wolters Kluwer; 2017. | ||
Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. An informational supplement, M100S. Wayne, PA: Clinical and Laboratory Standards Institute; 2016. | ||
Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62(2):316–322. | ||
Praharaj I, Sujatha S, Parija SC. Phenotypic & genotypic characterization of vancomycin resistant Enterococcus isolates from clinical specimens. Indian J Med Res. 2013;138(4):549–556. | ||
Biavasco F, Foglia G, Paoletti C, et al. VanA-type enterococci from humans, animals, and food: species distribution, population structure, Tn1546 typing and location, and virulence determinants. Appl Environ Microbiol. 2007;73(10):3307–3319. | ||
Oh JY, An S, Jin JS, Lee YC, Cho DT, Lee JC. Phenotypic and genotypic differences of the vancomycin-resistant Enterococcus faecium isolates from humans and poultry in Korea. J Microbiol. 2007;45(5):466–472. | ||
Sharifi Y, Hasani A, Ghotaslou R, et al. Survey of Virulence Determinants among Vancomycin Resistant Enterococcus faecalis and Enterococcus faecium Isolated from Clinical Specimens of Hospitalized Patients of North west of Iran. Open Microbiol J. 2012;6:34–39. | ||
Emaneini M, Aligholi M, Aminshahi M. Characterization of glycopeptides, aminoglycosides and macrolide resistance among Enterococcus faecalis and Enterococcus faecium isolates from hospitals in Tehran. Pol J Microbiol. 2008;57(2):173–178. | ||
Kafil HS, Asgharzadeh M. Vancomycin-resistant enteroccus faecium and enterococcus faecalis isolated from education hospital of iran. Maedica. 2014;9(4):323–327. | ||
Askarian M, Afkhamzadeh R, Monabbati A, Daxboeck F, Assadian O. Risk factors for rectal colonization with vancomycin-resistant enterococci in Shiraz, Iran. Int J Infect Dis. 2008;12(2):171–175. | ||
Yoon YK, Sim HS, Kim JY, et al. Epidemiology and control of an outbreak of vancomycin-resistant enterococci in the intensive care units. Yonsei Med J. 2009;50(5):637–643. | ||
Yang J, Jiang Y, Guo L, Ye L, Ma Y, Luo Y. Prevalence of Diverse Clones of Vancomycin-Resistant Enterococcus faecium ST78 in a Chinese Hospital. Microb Drug Resist. 2016;22(4):294–300. | ||
Sood S, Malhotra M, Das BK, Kapil A. Enterococcal infections & antimicrobial resistance. Indian J Med Res. 2008;128(2):111–121. | ||
Hirdon A, Edwards J, Patel J. Antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control. 2008;29(11):996–1011. | ||
Moaddab S, Kazemi-Haki B, Ebrahimi-Atashkhosroo N. Investigation of genes responsible for vancomycin resistance by Multiplex-PCR among enterococci isolated strains from inpatients and outpatients. Med Sci J. 2015;24(4):227–234. | ||
Shokoohizadeh L, Mobarez AM, Alebouyeh M, Zali MR, Ranjbar R. Genotyping of clinical and environmental multidrug resistant Enterococcus faecium strains. Indian J Pathol Microbiol. 2017;60(1):74–78. | ||
Karki S, Houston L, Land G, et al. Prevalence and risk factors for VRE colonisation in a tertiary hospital in Melbourne, Australia: a cross sectional study. Antimicrob Resist Infect Control. 2012;1(1):31. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.