Back to Journals » Infection and Drug Resistance » Volume 12
Molecular Detection Of Multidrug-Resistant Salmonella Isolated From Livestock Production Systems In South Africa
Authors Mthembu TP , Zishiri OT , El Zowalaty ME
Received 9 April 2019
Accepted for publication 12 July 2019
Published 14 November 2019 Volume 2019:12 Pages 3537—3548
DOI https://doi.org/10.2147/IDR.S211618
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eric Nulens
Thobeka P Mthembu,1 Oliver T Zishiri,1 Mohamed E El Zowalaty2,3
1School of Life Sciences, University of KwaZulu-Natal, Durban, 4000, South Africa; 2Virology and Microbiology Research Group, School of Health Sciences, University of KwaZulu-Natal, Durban, 4000, South Africa; 3Infectious Diseases and Anti-Infective Research Group, College of Pharmacy, University of Sharjah, Sharjah, 27272, UAE
Correspondence: Mohamed E El Zowalaty
Department of Pharmacy, City University College of Ajman, Ajman 18484, UAE
Email [email protected]
Background: Antibiotic-resistant bacterial pathogens associated with livestock remain a major concern worldwide as they get transmitted from animals to humans and cause foodborne and zoonotic diseases.
Methods: Antimicrobial resistance in livestock-associated Salmonella spp in South Africa was investigated using molecular DNA methods. Three hundred and sixty-one environmental faecal samples were randomly collected from avian (chicken and ducks), cows, pigs, goats, and sheep. Salmonella spp. were isolated on selective media and were confirmed using the polymerase chain reaction. Antimicrobial susceptibility testing against ampicillin, chloramphenicol, ciprofloxacin, ceftriaxone, azithromycin, tetracycline, amoxicillin-clavulanate and trimethoprim-sulfamethoxazole was determined using the Kirby–Bauer disk diffusion method. Isolates were screened for the presence of blaTEM-1, blaCMY-2, tetA, tetC, sul2 and dfrA7 resistance genes by PCR.
Results: Most of the isolates were resistant to ampicillin (64%), tetracycline (63%), amoxicillin-clavulanate (49%), trimethoprim-sulfamethoxazole (38%), and ceftriaxone (20%). Eight percent of the tested isolates were ciprofloxacin-resistant Salmonella spp. Multidrug resistance was observed with the mean multiple antibiotic resistance (MAR) index of 0.31. The study demonstrated that 43% of the isolates were multiple drug resistant. The prevalence rates of resistance genes were 44% for blaTEM-1, 35% for blaCMY-2, 21% for sul2, 18% for tetC, 14% for dfrA7 and 8% for tetA.
Conclusion: Resistance to ceftriaxone, detection of blaCMY-2 gene and the high level of intermediate susceptibility (33%) against ciprofloxacin suggested that livestock carry problematic Salmonella spp. This study used the global one-health initiative to report the potential public health risks of livestock-associated pathogens and highlights the importance of monitoring the trends of antimicrobial resistance for sustainability of antibiotics.
Keywords: Salmonella, food-borne pathogens, zoonotic, animals, antimicrobial, resistance, blaTEM-1, blaCMY-2, bacteria, ciprofloxacin
Introduction
The emergence of zoonotic bacterial infections and the ever-increasing occurrence of antimicrobial-resistant bacteria is a global concern.1 The adverse effects caused by antimicrobial-resistant bacteria in both humans and domestic animals draw significant attention to these pathogens and pose a need for them to be monitored to ensure human biosecurity.2–4 Antimicrobial-resistant bacteria decrease the efficiency of available antibiotics, resulting in infection treatment failures which sometimes lead to high morbidity and mortality.5 Salmonella is a zoonotic bacterial pathogen that is responsible for gastroenteritis, focal infection, enteric fever (typhoid) and bacteremia in humans.6–8 Enteric fever is caused by Salmonella typhi and its subtypes, which infect only humans while the other diseases are caused by the non-typhoidal Salmonella.7 Gastroenteritis (or food poisoning) is the most common disease caused by non-typhoidal Salmonella, which is normally treated by fluid supplementation.9 Antimicrobial agents are recommended only in severe cases in children under the age of 5, elderly people (over 65 years) and immunocompromised individuals.5 Antibiotics are categorized according to their mode of action in bacteria. Depending on the type of the antimicrobial agent, the activities of the antibiotics result in bacterial cell wall synthesis disruption, inhibition of nucleic acids synthesis, inhibition of protein synthesis or folic acid pathway inhibition.10,11 The use of antibiotics for treatment of salmonellosis is challenged by the continuous emergence of antimicrobial-resistant strains.12
The antibiotic resistance challenge dates back to the 1960s when antimicrobial-resistant Salmonella typhimurium was reported.13 Ampicillin (beta-lactam), sulfamethoxazole (sulfonamide), furazolidone (nitrofuran) and tetracyclines were reported to have decreased efficiency against S. typhimurium treatment in calves and humans.14 Resistance to antibiotics such as gentamicin, kanamycin, streptomycin, chloramphenicol and trimethoprim was detected in the subsequent years.15–17 In the late 1990s, resistance to nalidixic acid (fluoroquinolone) and ceftriaxone (third-generation cephalosporin) was reported following an outbreak in cattle and humans in the USA.14,18 Antibiotic-resistant Salmonella spp. were reported to newer drugs such as azithromycin and ciprofloxacin.19,20 In addition, fluoroquinolone-resistant Salmonella spp. are listed among the high priority list of the World Health Organization (WHO).21,22 The major concern is that once a drug is discovered it takes a very short period of time for the bacteria to develop resistance to it as a result of quick response to environmental stimuli, short generation time by bacteria and horizontal sharing of resistance genes between bacterial species.23,24 On the other hand, it takes a many years for a new antibiotic to be developed, and approved for clinical use.25–28 Antibiotic resistance is mainly attributed to the improper and injudicious use of antibiotics as growth promoters in animal feeds as well as during treatment and for prophylactic purposes.29,30
Adminstration of small doses of antibiotics to healthy animals causes the microbiota in the guts of animals to be familiar with the drugs and gives them a chance to develop resistance towards the given substances.19,30,31 The bacteria then share the antibiotic resistance determinants via mobile genetic elements (MGEs) such as plasmids, transposons, integrons and phages.32,33 This renders the healthy animals as carriers of antibiotic-resistant bacteria.34 Since antibiotic-resistant bacteria are found in the gut, faecal contamination is the main route of transmission of these pathogens to humans. The environments in which the animals are reared and direct contact with animals are possible sources of transmission of resistant zoonotic pathogens to humans.35,36
The mechanisms encoded by antimicrobial resistance determinants include antimicrobial modification and inactivation, alteration of the antimicrobial target site, efflux pumps and membrane impermeability.37,38 These protect the bacteria from being attacked by antibiotics.37 Bacteria which are resistant to the beta-lactam class of antibiotics produce beta-lactamase enzyme which destroys the beta-lactam ring thus deactivating the antibiotic.10,23 Macrolide repressor protein produced by Salmonella spp. inactivates azithromycin and erythromycin.39 Resistance to quinolones is achieved by mutations in DNA gyrase coding genes (gyrA and gyrB) thus modifying the target site of the antibiotic.40 Efflux pumps transport substances out of the cell, including detergents, dyes and antibiotics.41 Salmonella spp. uses the well-studied AcrAB-TolC efflux pump to extrude antibiotics such as tetracycline, chloramphenicol and quinolones.42,43 Bacteria resist antibiotic entry into the cell by reducing or modifying porin channels in the outer membrane which are used by antibiotic molecules to enter the bacterial cell to reach their targets.44,45 Genes which encode for antibiotic resistance are either located on the chromosome or plasmids within a bacterial cell and are mobilized by transposons and integrons during conjugation or phages through transduction.33,46
Antibiotics used for the treatment of salmonellosis include ampicillin, tetracycline, amoxicillin-clavulanate, chloramphenicol, trimethoprim-sulfamethoxazole, ciprofloxacin, ceftriaxone and azithromycin.47,48 Salmonella spp. utilize several resistance mechanisms to escape the antimicrobial activities. The alarming rates of antimicrobial resistance led to the investigation of the genetic determinants in bacteria. Several studies focused on the detection of antimicrobial resistance genes which are the drivers of antimicrobial resistance.5,37,40
The use of antibiotics for growth promotion in livestock decreases their efficiency and therefore the effectiveness of the antibiotics differs between countries depending on the available antibiotic regulatory practices.30,49 It was reported that a decrease in resistance to antibiotics such as tylosin, which were banned for use as growth promoters in European countries while resistance rates are still rising in countries where this antibiotic is used as a growth promoter.50 Screening bacterial pathogens for the presence of antibiotic resistance genes and detection by molecular methods enable researchers to determine whether a drug will be effective in a certain area. This aids in appropriate prescription of the correct treatment to patients with a particular bacterial infection, rather than prescription of an antibiotic which may result in the development of antimicrobial resistance.4 Against this background, the current study aimed to determine the antimicrobial susceptibility profiles and to detect resistance determinants of Salmonella spp. isolated from livestock production systems in South Africa.
Materials And Methods
Ethical Approval
The study was approved by the Animal Research Ethics Committee of the University of Kwa-Zulu Natal (Reference numbers AREC/051/017M, AREC 071/017 and AREC 014/018). The field sampling protocols, samples collected from animals, and the research were conducted in full compliance with Section 20 of the Animal Diseases Act of 1984 (Act No 35 of 1984) and were approved by the South African Department of Agriculture, Forestry and Fisheries DAFF (Section 20 approval reference number 12/11/1/5 granted to Prof ME El Zowalaty).
Samples And Pre-Enrichment
A total of three hundred and sixty-one (361) faecal and environmental samples were collected from different animal hosts including cattle, sheep, goats, pigs, ducks and chickens which were housed in small-scale commercial farms in Flagstaff (O.R Tambo, Eastern Cape), Verulam (eThekwini, KwaZulu-Natal) and South Coast (Amandawe and Mtwalume, UGU, KwaZulu-Natal) as depicted in Figure 1. All samples as shown in Table 1 were randomly collected between May and August 2018. In the farms, livestock are bought from large-scale farms and sold to the local communities. Oral and rectal swab samples were collected from animals using sterile collection swabs and transferred into 10 mL of 0.1% buffered sterilized peptone water (Merck, South Africa). Fresh environmental faecal samples emanating from purportedly healthy livestock, as well as samples from their environments including soil, feed and water, were also randomly collected using sterile collection swabs. All swab samples were transferred into 10 mL of 0.1% buffered sterilized peptone water (Merck, South Africa). Water samples were collected from the containers inside the livestock housing. Samples were transported on ice to the discipline of genetics laboratories at University of KwaZulu-Natal Westville campus where enrichment of the samples was done by incubating the swabs in 10 mL of 0.1% buffered peptone water at 37°C overnight.
 |
Table 1 Number And Type Of Samples Collected From Farms In Flagstaff, Verulam And South Coast In South Africa |
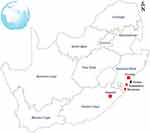 |
Figure 1 Map of South Africa showing the geographic locations of the farms where the samples used in this study were collected. |
Selective Enrichment
From each of the enriched samples, 0.1 mL was aseptically transferred into 10 mL of Rappaport Vassiliadis (RV) broth (Sigma-Aldrich, India) and incubated for 24 hrs at 42°C. RV is a selective medium that is enriched with malachite green which inhibits the growth of microorganisms other than Salmonella. A previously identified and confirmed Salmonella enterica was used as a positive control.51 Bacterial isolation was performed on Xylose-Lysine-Deoxycholate (XLD) agar (Sigma-Aldrich, Switzerland) by aseptically streaking a loopful of the culture from RV broth onto the plates. S. enterica is differentiated from Escherichia coli and Shigella spp. by producing red colonies with black centers on XLD agar. After 24 hrs of incubation at 35°C, the plates were observed for the growth of the expected colonies. Representative single colonies of the correct morphology were randomly picked from each plate and transferred into tubes containing 10 mL of tryptose soy broth (Merck, South Africa) and incubated at 37°C for 18–24 hrs. A 2 mL of the culture was used for DNA extraction. Equal amounts of 0.5 mL each of 60% glycerol and Salmonella pure culture were mixed in 1.5 mL cryotubes and stored at −80°C for future use.
DNA Extraction
Total genomic DNA was extracted from Salmonella cultures using the conventional boiling method. One milliliter of the cultured sample was transferred into 1.5 mL Eppendorf tube and centrifuged at 14,000 × g for 5 mins. The supernatant was discarded and another 1 mL of culture was added to the pellet and centrifuged again to get a larger pellet. Six hundred µL of sterile distilled water was added to the pellet and centrifuged for 5 mins at 14,000 × g. The supernatant was discarded, 200 µL of sterile distilled water was added again and incubated in a heating block at 100°C (Labnet, USA). The sample was boiled for 10 mins with immediate cooling on ice for 5 mins. After cooling, the sample was centrifuged at 14,000 × g for 5 mins; the resulting supernatant was transferred into a fresh Eppendorf tube and stored at −20°C for further use.
Molecular Confirmation Of Salmonella spp. Using PCR
Salmonella spp. were confirmed by amplifying the invA gene, which is genus-specific gene and the iroB gene for identification of S. enterica using specific primers as previously reported52,53 as shown in Table 2. The PCR reaction volume was 25 µL which consisted of 12.5 µL Dream Taq master mix (Thermo-Fischer Scientific, South Africa), 0.5 µL each of forward and reverse primers, 6.5 µL sterile distilled water and 5 µL template DNA. The PCR reaction conditions consisted of 34 amplification cycles for invA gene using the following conditions: denaturation for 30 s at 95°C, annealing for 30 s at 58°C, extension for 1 min at 72°C and final extension for 5 mins at 72°C. The same amplification parameters were used for the iroB gene using a different annealing temperature of 58°C. The PCR amplicons were stored at −20°C till future use.
 |
Table 2 Forward And Reverse Primer Sequences Of invA And iroB Genes |
Antimicrobial Susceptibility Testing
Salmonella spp. isolates were cultured into 10 mL of tryptone soy broth at 37°C for 18 hrs. Isolates were tested for their antimicrobial susceptibility profiles according to the CLSI guidelines.54 Antibiotic disks containing ampicillin (10 µg), trimethoprim and sulfamethoxazole (25 µg), amoxicillin-clavulanate (20 µg), chloramphenicol (30 µg), ciprofloxacin (5 µg), tetracycline (30 µg), ceftriaxone (30 µg) and azithromycin (15 µg) were placed on the inoculated Mueller-Hinton agar plates. E. coli ATCC 25922 reference strain was used as a positive control as recommended in the CLSI guidelines. The plates were incubated at 37°C for 16 hrs to examine susceptibility of the isolates to the antibiotics. The zone of inhibition (in millimeters) were determined by measuring the diameter of the area of no bacterial growth around the disk. The results were interpreted as susceptible, intermediate or resistant in compliance with CLSI recommendations.
PCR Amplification Of Antibiotic Resistance Genes
Screening of antimicrobial resistance genes in Salmonella isolates was performed by PCR methods using previously reported primers as shown in Table 3. The reaction tube contained 25 µL which was made up of 12.5 µL Dream Taq master mix (Thermo-Fischer Scientific, South Africa), 0.5 µL each of forward and reverse primers, 6.5 µL sterile distilled water and 5 µL template DNA. The negative control had sterile distilled water instead of DNA. E. coli ATCC 25922 reference strain was used as a positive control for some of the investigated resistance genes and previously extracted and known DNA from a pure S. enterica culture was used as positive control. PCR was conducted in a thermocycler (BioRad, Singapore) using the following conditions consisting of an initial denaturation for 3 mins at 95°C, followed by 30 amplification cycles of denaturation for 30 s at 95°C, annealing for 30 s at a specific temperature for each set of primers, extension for 1 min at 72°C and final extension for 8 mins at 72°C.
 |
Table 3 Primers And Annealing Temperatures For Antimicrobial Resistance Genes Amplification |
Statistical Analysis
Multiple antibiotic resistance (MAR) indices were determined to assess the risk level of the isolates using a previously reported method.60 The number of antibiotics that each isolate was resistant to was divided by the total number of antibiotics tested to determine the MAR index. Indices greater than 0.2 were regarded as high risk, meaning that the isolates were multidrug resistant and representing an origin of the isolates from a source of high use of antibiotics as was reported previously.60 Furthermore, inferential statistics (Fisher’s exact and correlation tests) using IBM SPSS version 25 and Microsoft excel 2010 were used to analyze the results. The Fischer’s exact test was implemented to investigate whether there was a relationship between antimicrobial resistance genes with animal host and farm location (area where samples were collected). The magnitude and direction of the correlation between antimicrobial resistance genes were evaluated using Pearson’s correlation test. Furthermore, the effect of farm location as a predictor of the presence of the genes in the present study was investigated by binary logistic regression. South coast was used as a reference category for comparing Flagstaff and Verulam. The statistical models evaluated the null hypothesis that there was no association between location, animal host and the presence of the antimicrobial resistance genes. The critical value of 0.05 was used to decide whether to reject or not reject the null hypothesis.
Results
Out of 361 samples, 195 (54%) showed positive growth on RV medium and XLD agar, this was observed by cloudiness of the RV broth after 24 hrs growth on XLD agar plates. The positive control showed red colonies with black centers, as expected for S. enterica on XLD agar. Out of 195 presumptive Salmonella spp., 106 were confirmed to be positive by the invA gene amplification (284 bp). S. enterica is responsible for infections in humans, with serovars such as Enteritidis and Typhimurium being the most reported.6 In this study, 30.2% of the Salmonella isolates were confirmed to be S. enterica by iroB gene amplification (606 bp). The 106 Salmonella isolates that were confirmed by invA gene amplification were further analyzed for antimicrobial susceptibility profiling and detection of resistance genes.
The results from Kirby–Bauer disk diffusion method demonstrated that the Salmonella isolates in this study were resistant to all the tested antibiotics (Figure 2). The prevalence of resistance was high to ampicillin (64%) followed by tetracycline (63%), amoxicillin-clavulanate (49%), trimethoprim-sulfamethoxazole (38%) and ceftriaxone (20%). Resistances to ciprofloxacin, chloramphenicol and azithromycin were low with proportions of 8%, 8% and 5%, respectively. It was found that 33% of the isolates were intermediate-resistant to ciprofloxacin. As illustrated in Figure 2, resistance in Salmonella spp. isolated was high in AMP, TE, AMC, SXT and CRO. The mean of MAR index was 0.31 (range 0–0.875) with 43% (n=46 out of 106) of the isolates displaying resistance to three or more antibiotics and were considered multidrug resistant. It was found that one isolate was resistant to seven antibiotics, while five, seven, 15 and 18 isolates were resistant to six, five, four, and three antibiotics out of the eight tested drugs. The multidrug resistance patterns are depicted in Table 4.
 |
Table 4 Multidrug Resistance Patterns Of Salmonella Isolates Resistant To Two Or More Antibiotics |
 |
Figure 2 The number of samples that were susceptible, intermediate or resistant to the tested antibiotics. |
All tested isolates collected from cows, pigs, sheep, and goat, and chickens possessed at least five of the six screened antimicrobial resistance genes using PCR except for isolates collected from ducks which had only four genes. Most of the AMR genes were found in Salmonella isolated from chicken as shown in Figure 3. The prevalence of the genes was as follows: blaTEM-1 (44%), blaCMY-2 (35%), sul2 (21%), tetC (18%), dfrA7 (14%) and tetA (8%).
 |
Figure 3 Distribution of antimicrobial resistance genes in Salmonella spp. isolated from livestock. |
There was no statistically significant (p>0.05) association between sample material and the animal host in the presence of antimicrobial resistance genes. However, farm location was significantly (p<0.05) associated with the presence of tetC, dfrA7 and blaTEM-1 as shown in Table 5.
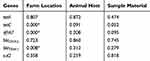 |
Table 5 Fisher’s Exact Test Results Showing Whether There Is A Significant Association (p-Values) Between The Antimicrobial Resistance Genes And Location, Animal Host As Well As Sample Material |
A significant (p<0.05) positive correlation was observed between tetC and blaCMY-2, moreover, blaCMY-2 was positively correlated to blaTEM-1 and sul2. The sul2 gene was also positively correlated to dfrA7 and blaTEM-1 as the p-values were less than the critical values of 0.05 and 0.01 (Table 6).
 |
Table 6 The Relationship Between The Detected Antimicrobial Resistance Genes As Statistically Assessed By Pearson’s Correlation |
The effect of farm location as a predictor for the presence of antimicrobial resistance genes was further assessed by binary logistic regression. Although the majority of the results were not significant (p>0.05) it was observed that the presence of genes in the South coast was more related to Flagstaff than Verulam with regard to the odds ratios (Table 7). Higher odds ratios were observed when comparing South coast to Flagstaff than when comparing South coast to Verulam. The effect of farm location as a predictor of the presence of dfrA7 was significant (p<0.05) when Verulam was compared to South coast and the effect on the presence of blaTEM-1 was significant (p<0.05) when South Coast was compared to Verulam as shown in Table 7. The presence of tetC was significant (p<0.05) when all the three farm locations were compared.
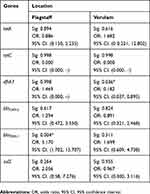 |
Table 7 The Effect Of Farm Location As A Predictor Of The Presence Of The Investigated Antimicrobial Resistance Genes |
Discussion
The findings of the current study indicate the presence of multidrug-resistant Salmonella spp. isolated from livestock in South Africa. High rates of resistance were observed against ampicillin, tetracycline, amoxicillin-clavulanate, trimethoprim-sulfamethoxazole and ceftriaxone with frequencies of 64%, 63%, 49%, 38% and 20%, respectively. There was also decreased activity against ciprofloxacin, chloramphenicol and azithromycin as some of the isolates were also resistant to these antibiotics (Figure 2). These high resistance rates detected in livestock-associated Salmonella spp. in South Africa may be explained that antibiotics use as growth-promoting agents is legally allowed in the country.61 In addition, imprudent use of antibiotics in livestock farms was recently reported in South Africa.62
The findings in the current study were in line with other studies which reported high resistance rates towards ampicillin, tetracycline, trimethoprim-sulfamethoxazole.63–65 Higher resistance rates against azithromycin (16.9%) and ceftriaxone (24.07%) were previously reported66 as compared to the current study. A decrease in the efficiency of chloramphenicol and ciprofloxacin was previously reported with intermediate susceptibilities of 67% and 33%, respectively.67 Tetracycline, ampicillin and amoxicillin are commonly used for the treatment of infections in cattle, pigs, poultry, sheep and goats.68 During treatment, antibiotics are administered to all livestock herd on the farm thus triggering antimicrobial resistance and affects the intestinal microflora of healthy animals. Other than disease treatment and prevention, antibiotics such as tetracyclines, sulfonamides, macrolides, fluoroquinolones and beta-lactams are mainly used as growth promoters in farms.30,50,69 This can be an explanation for the high rates of resistance towards these antibiotics. Even though the resistance frequencies were low for ciprofloxacin, chloramphenicol and azithromycin, the high level of intermediate susceptibility towards ciprofloxacin, is concerning as it indicates the possibility of resistance towards this antibiotic in the future. The multidrug resistance phenotype displayed by 43% of the isolates reveals that humans in proximity to these farms are at risk of serious zoonotic infections with difficult-to-treat Salmonella spp. The occurrence of antimicrobial-resistant Salmonella spp. in this study indicates that food animals and their environments are the source of antimicrobial resistance.
Antibiotic resistance is mediated by genetic elements which code for different mechanisms which are used by the bacteria to escape the effects of antibiotics.37 Beta-lactam resistance genes (blaTEM-1, blaCMY-2), sulfonamide resistance gene (sul2), tetracycline resistance genes (tetA and tetC) and trimethoprim resistance determinant (dfrA7) were observed in this study. The blaTEM-1 and blaCMY-2 genes code for enzymes which degrade beta-lactam antibiotics by hydrolyzing the beta-lactam ring.10,70 The sul2 and dfrA7 genes are involved in synthesizing the insensitive forms of dihydropteroate and dihydrofolate reductase to escape the activity of trimethoprim-sulfamethoxazole, respectively.71 TetC and tetA are responsible for transporting tetracycline out of the cell.72 The antimicrobial resistance genes were distributed in all livestock animals; however, higher prevalence was observed in chickens (Figure 3). Chickens are reared in large numbers in closed environments thus creating favourable conditions for pathogens and disease dissemination leading to frequent treatment. Poultry production systems such as battery cages, litter systems and automated feeders also promote antimicrobial resistance dissemination in industrialized farms.73 Similar studies which screened antimicrobial resistance genes from food animals reported lower rates of blaTEM-1 of 19.3%63 and 24.73%74 as compared to the current study. Very low prevalence of tetA gene was previously reported.51,67 The prevalence of blaCMY-2 in the current study was similar to a previous study which reported a rate of 38.88%.75 The high prevalence of beta-lactam degrading genetic determinants in this study is of concern because extended beta-lactams such as ceftriaxone are the drug of choice for salmonellosis treatment in children and pregnant women.42
The prevalence of tetC, dfrA7 and blaTEM-1 genes was significantly (p<0.05) associated with farm location (Table 5). These genes were highly prevalent in Verulam, which is more urbanized as compared to South coast and Flagstaff (Table 7). A possible explanation is that livestock in urban areas are mostly fed on preserved foods with traces of chemicals and antibiotics while in rural areas livestock are normally fed on natural grass and grains. As a result of high demand for meat, over-population and limited space, food animals in urban areas are commonly fed antibiotics for fast growth.29,76 The prevalence of tetC, dfrA7 and blaTEM-1 suggested that tetracycline, trimethoprim and ampicillin are frequently used on the farms in this study, particularly Verulam.
There was a significant (p<0.05) weak positive correlation (ranging from 0.34 to 0.19) between tetC, blaCMY-2, blaTEM-1, sul2 and dfrA7 (Table 6). The roles of these genes and their locations in Salmonella could explain their correlation. The sul2 and dfrA7 genes are cassettes found in class 1 integrons which are associated with antimicrobial resistance and are responsible for biosynthetic processes in Salmonella.59 The tetC gene and blaCMY-2 are related by their involvement in cell wall synthesis and membrane transport activity.10,43 The blaTEM-1 and blaCMY-2 genes have the same activity of degrading beta-lactams and extended beta-lactams which could explain their association.10 Furthermore, blaTEM-1 and blaCMY-2 are located on transferable plasmids.70 The association between sul2 and beta-lactamase-producing genes (blaTEM-1 and blaCMY-2) can be explained by their location in Salmonella.37
The associations between resistance genes in the current study are in accordance with the previously reported results where it was found that class1 integrons are related to transposons and insertion sequence (IS) elements which carry antimicrobial resistance genes.77 Resistance genes are usually carried by integrons as gene cassettes.33 The dfrA7 gene is one of the classic genes carried by the class 1 integron.78 The sul2 gene is also found in the class 1 integron which further explains the relationship between these genes. Beta-lactamase genes (blaTEM-1 and blaCMY-2) are located in the IS elements which are linked to the class 1 integrons77 which explained the correlation between these genes in the current study.
High levels of antimicrobial resistance phenotypes were observed compared to the resistance genetic determinants. Highlighted in this study is the risk that humans are exposed to as a result of livestock-associated antibiotic-resistant Salmonella. A high level of intermediate susceptibility towards ciprofloxacin provokes questions about the efficiency of this antibiotic in future. In addition, in the current study the detection of fluoroquinolone-resistant Salmonella spp. which is listed in the WHO high priority list22 is very concerning. The presence of blaCMY-2 gene which confers Salmonella resistance to cephalosporins (ceftriaxone) is also a matter of concern as this antibiotic is one of the lately introduced antibiotics for medicinal use. The beta-lactam class of antibiotics is very important for treatment of salmonellosis in children, who are at risk of being infected. Another matter to point out is that these antibiotics are not only used to treat salmonellosis, but also infections by other bacterial pathogens.
The lack of human samples and the limited number of screened samples are among the limitations in the present study, which can be used to draw conclusions on the spread of Salmonella from livestock to humans. Typing the Salmonella isolates to the serovar level in order to determine the dominant Salmonella serovars in farms and human samples in South Africa require further investigation. Whole-genome sequence of selected samples will provide in-depth genomic characterization of resistome and virulome of the isolates. Frequent monitoring and enhanced surveillance programs will serve as an early warning system of antibiotic-resistant Salmonella, helping us find any potential disease much more quickly, control antibiotic resistance at the farm level, and minimize the public health burden.
Conclusions
The results of this study indicate the importance of monitoring the effects of antimicrobial use in livestock to minimize the public health risks. The findings of this study highlights the improtance of monitoring the trend of antimicrobial resistance in South Africa and will contribute to the global One Health initiative to contain antimicrobial resistance.
Acknowledgments
We would like to thank the four anonymous farm owners who allowed us to collect the samples for this research project from their farms. We would like to thank the South African National Research Foundation for supporting this research through the Thuthuka Funding Instrument (grant number TTK170411226583). We would also like to thank the College of Agriculture, Engineering and Science as well as the School of Life Sciences at University of KwaZulu-Natal (Westville campus) for their support in numerous ways during the execution of this research.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Robinson TP, Bu D, Carrique-Mas J, et al. Antibiotic resistance is the quintessential One Health issue. Trans R Soc Trop Med Hyg. 2016;110(7):377–380. doi:10.1093/trstmh/trw048
2. Friedman ND, Temkin E, Carmeli Y. The negative impact of antibiotic resistance. Clin Microbiol Infect. 2016;22(5):416–422. doi:10.1016/j.cmi.2015.12.002
3. Michael CA, Dominey-Howes D, Labbate M. The antimicrobial resistance crisis: causes, consequences, and management. Front Public Health. 2014;2:145. doi:10.3389/fpubh.2014.00145
4. Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. Pharm Therapeut. 2015;40(4):277.
5. Eng S-K, Pusparajah P, Ab Mutalib N-S, Ser H-L, Chan K-G, Lee L-H. Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci. 2015;8(3):284–293. doi:10.1080/21553769.2015.1051243
6. Andino A, Hanning I. Salmonella enterica: survival, colonization, and virulence differences among serovars. Sci World J. 2015;2015. doi:10.1155/2015/520179
7. Chen H-M, Wang Y, Su L-H, Chiu C-H. Nontyphoid Salmonella infection: microbiology, clinical features, and antimicrobial therapy. Pediatr Neonatol. 2013;54(3):147–152. doi:10.1016/j.pedneo.2013.01.010
8. Uche IV, MacLennan CA, Saul A. A systematic review of the incidence, risk factors and case fatality rates of invasive nontyphoidal Salmonella (iNTS) disease in Africa (1966 to 2014). PLoS Negl Trop Dis. 2017;11(1):e0005118. doi:10.1371/journal.pntd.0005118
9. Li Y, Xie X, Xu X, et al. Nontyphoidal Salmonella infection in children with acute gastroenteritis: prevalence, serotypes, and antimicrobial resistance in Shanghai, China. Foodborne Pathog Dis. 2014;11(3):200–206. doi:10.1089/fpd.2013.1629
10. Bush K, Bradford PA. β-Lactams and β-lactamase inhibitors: an overview. Cold Spring Harb Perspect Med. 2016;6(8):a025247. doi:10.1101/cshperspect.a025247
11. Redgrave LS, Sutton SB, Webber MA, Piddock LJ. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014;22(8):377–380. doi:10.1016/j.tim.2014.04.007
12. Allcock S, Young E, Holmes M, et al. Antimicrobial resistance in human populations: challenges and opportunities. Glob Health Epidemiol Genom. 2017;2.
13. Threlfall EJ. Epidemic Salmonella typhimurium DT 104—a truly international multiresistant clone. J Antimicrob Chemother. 2000;46(1):7–10. doi:10.1093/jac/46.1.7
14. Threlfall EJ. Antimicrobial drug resistance in Salmonella: problems and perspectives in food-and water-borne infections. FEMS Microbiol Rev. 2002;26(2):141–148. doi:10.1111/j.1574-6976.2002.tb00606.x
15. Chaslus-Dancla E, Martel J-L, Carlier C, Lafont J, Courvalin P. Emergence of aminoglycoside 3-N-acetyltransferase IV in Escherichia coli and Salmonella typhimurium isolated from animals in France. Antimicrob Agents Chemother. 1986;29(2):239–243. doi:10.1128/aac.29.2.239
16. Neu HC. The crisis in antibiotic resistance. Science. 1992;257(5073):1064–1073. doi:10.1126/science.257.5073.1064
17. Threlfall E, Rowe B, Ward L. A comparison of multiple drug resistance in salmonellas from humans and food animals in England and Wales, 1981 and 1990. Epidemiol Infect. 1993;111(2):189–198. doi:10.1017/S0950268800056892
18. Rowe B, Ward LR, Threlfall EJ. Multidrug-resistant Salmonella typhi: a worldwide epidemic. Clin Infect Dis. 1997;24(Supplement_1):S106–S109. doi:10.1093/clinids/24.supplement_1.s106
19. Cui S, Li J, Sun Z, et al. Ciprofloxacin-resistant Salmonella enterica serotype Typhimurium, China. Emerg Infect Dis. 2008;14(3):493. doi:10.3201/eid1403.070857
20. Wang J, Li Y, Xu X, et al. Antimicrobial resistance of Salmonella enterica serovar Typhimurium in Shanghai, China. Front Microbiol. 2017;8:510.
21. Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi:10.1016/S1473-3099(17)30753-3
22. Tacconelli E, Magrini N, Kahlmeter G, Singh N. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organ. 2017;27.
23. Aminov RI. The role of antibiotics and antibiotic resistance in nature. Environ Microbiol. 2009;11(12):2970–2988. doi:10.1111/j.1462-2920.2009.01972.x
24. Deris JB, Kim M, Zhang Z, et al. The innate growth bistability and fitness landscapes of antibiotic-resistant bacteria. Science. 2013;342(6162):1237435. doi:10.1126/science.1237435
25. Aminov RI. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol. 2010;1:134. doi:10.3389/fmicb.2010.00143
26. Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645. doi:10.2147/IDR.S173867
27. Ferri M, Ranucci E, Romagnoli P, Giaccone V. Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr. 2017;57(13):2857–2876. doi:10.1080/10408398.2015.1077192
28. Gwynn MN, Portnoy A, Rittenhouse SF, Payne DJ. Challenges of antibacterial discovery revisited. Ann N Y Acad Sci. 2010;1213(1):5–19. doi:10.1111/j.1749-6632.2010.05828.x
29. Chattopadhyay MK. Use of antibiotics as feed additives: a burning question. Front Microbiol. 2014;5:334. doi:10.3389/fmicb.2014.00547
30. Eagar H, Swan G, Van Vuuren M. A survey of antimicrobial usage in animals in South Africa with specific reference to food animals. J S Afr Vet Assoc. 2012;83(1):15–23. doi:10.4102/jsava.v83i1.16
31. van Vuuren M. Bacterial resistance against antibiotics: global and local trends. Stockfarm. 2017;7(5):51.
32. Gillings MR. Integrons: past, present, and future. Microbiol Mol Biol Rev. 2014;78(2):257–277. doi:10.1128/MMBR.00056-13
33. Hall RM. Integrons and gene cassettes: hotspots of diversity in bacterial genomes. Ann N Y Acad Sci. 2012;1267(1):71–78. doi:10.1111/j.1749-6632.2012.06588.x
34. Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev. 2011;24(4):718–733. doi:10.1128/CMR.00002-11
35. Dhama K, Rajagunalan S, Chakraborty S, et al. Food-borne pathogens of animal origin-diagnosis, prevention, control and their zoonotic significance: a review. Pak J Biol Sci. 2013;16(20):1076. doi:10.3923/pjbs.2013.1076.1085
36. Tauxe RV. Emerging foodborne pathogens. Int J Food Microbiol. 2002;78(1–2):31–41. doi:10.1016/s0168-1605(02)00232-5
37. Frye JG, Jackson CR. Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. isolated from US food animals. Front Microbiol. 2013;4:135. doi:10.3389/fmicb.2013.00077
38. Kapoor G, Saigal S, Elongavan A. Action and resistance mechanisms of antibiotics: a guide for clinicians. J Anaesthesiol Clin Pharmacol. 2017;33(3):300. doi:10.4103/joacp.JOACP_349_15
39. Nair S, Ashton P, Doumith M, et al. WGS for surveillance of antimicrobial resistance: a pilot study to detect the prevalence and mechanism of resistance to azithromycin in a UK population of non-typhoidal Salmonella. J Antimicrob Chemother. 2016;71(12):3400–3408. doi:10.1093/jac/dkw318
40. McDermott PF, Tyson GH, Kabera C, et al. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother. 2016;60(9):5515–5520. doi:10.1128/AAC.01030-16
41. Baugh S, Ekanayaka AS, Piddock LJ, Webber MA. Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J Antimicrob Chemother. 2012;67(10):2409–2417. doi:10.1093/jac/dks228
42. Parry CM, Threlfall E. Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr Opin Infect Dis. 2008;21(5):531–538. doi:10.1097/QCO.0b013e32830f453a
43. Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem Biophys Res Commun. 2014;453(2):254–267. doi:10.1016/j.bbrc.2014.05.090
44. Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta. 2009;1794(5):808–816. doi:10.1016/j.bbapap.2008.11.005
45. van der Heijden J, Reynolds LA, Deng W, et al. Salmonella rapidly regulates membrane permeability to survive oxidative stress. MBio. 2016;7(4):e01238–e01216. doi:10.1128/mBio.01238-16
46. Guerra B, Soto S, Helmuth R, Mendoza MC. Characterization of a self-transferable plasmid from Salmonella enterica serotype Typhimurium clinical isolates carrying two integron-borne gene cassettes together with virulence and drug resistance genes. Antimicrob Agents Chemother. 2002;46(9):2977–2981. doi:10.1128/aac.46.9.2977-2981.2002
47. Crump JA, Medalla FM, Joyce KW, et al. Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob Agents Chemother. 2011;55(3):1148–1154. doi:10.1128/AAC.01333-10
48. Sjölund-Karlsson M, Joyce K, Blickenstaff K, et al. Antimicrobial susceptibility to azithromycin among Salmonella enterica isolates from the United States. Antimicrob Agents Chemother. 2011;55(9):3985–3989. doi:10.1128/AAC.00590-11
49. Alonso C, Zarazaga M, Ben Sallem R, Jouini A, Ben Slama K, Torres C. Antibiotic resistance in Escherichia coli in husbandry animals: the African perspective. Lett Appl Microbiol. 2017;64(5):318–334. doi:10.1111/lam.12724
50. Van Boeckel TP, Brower C, Gilbert M, et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci. 2015;112(18):5649–5654. doi:10.1073/pnas.1503141112
51. Zishiri OT, Mkhize N, Mukaratirwa S. Prevalence of virulence and antimicrobial resistance genes in Salmonella spp. isolated from commercial chickens and human clinical isolates from South Africa and Brazil. Onderstepoort J Vet Res. 2016;83(1):1–11. doi:10.4102/ojvr.v83i1.1067
52. Li, H., Bhaskara, A., Megalis, C. & Tortorello, M.L., 2012. Transcriptome analysis of Salmonella desiccation resistance’, Foodborne Pathogens and Disease.2012;12;1143–1151. doi:10.1089/fpd.2012.1254.
53. Bäumler AJ, Heffron F, Reissbrodt R. Rapid detection of Salmonella enterica with primers specific for iroB. J Clin Microbiol. 1997;35(5):1224–1230.
54. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing of Anaerobic Bacteria: Informational Supplement. Clinical and Laboratory Standards Institute (CLSI), Wayne, PA, USA; 2009.
55. Poppe C, Martin LC, Gyles CL, Reid-Smith R, Boerlin P, McEwen SA, et al.Acquisition of resistance to extended-spectrum cephalosporins by Salmonella enterica subsp. enterica serovar Newport and Escherichia coli in turkey poult intestinal tract. Appl Environ Microbiol. 2005;71(3):1184–9210.1128.
56. Guerra B, Soto SM, Argüelles JM, Mendoza MC. Multidrug resistance is mediated by large plasmids carrying a class 1 integron in the emergent Salmonella enterica serotype [4, 5, 12: i:−]. Antimicrobial agents and chemotherapy. 2001;45(4):1305–1308.
57. Fonseca EL, Mykytczuk OL, Asensi MD, Reis EM, Ferraz LR, Paula FL, Ng LK, Rodrigues DP. Clonality and antimicrobial resistance gene profiles of multidrug-resistant Salmonella enterica serovar Infantis isolates from four public hospitals in Rio de Janeiro, Brazil. Journal of clinical microbiology. 2006;44(8):2767-2772.
58. Poppe CL, Martin A, Muckle MA, McEwen S, Weir E. Characterization of antimicrobial resistance of Salmonella Newport isolated from animals, the environment, and animal food products in Canada. Can J Vet Res. 2006;70:105–114.
59. Frank T, Gautier V, Talarmin A, Bercion R, Arlet G. Characterization of sulphonamide resistance genes and class 1 integron gene cassettes in Enterobacteriaceae, Central African Republic (CAR). J Antimicrob Chemother. 2007;59(4):742–745. doi:10.1093/jac/dkl538
60. Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46(1):165–170.
61. Department of Agriculture FaF, South Africa (DAFF). Fertilizer, Farm Feeds, Agricultural Remedies and Stock Remedies Act, 1947; Publication of Farm Feeds (animal Feeds) Policy for Public Comments. Act No. 36 of 1947 C.F.R. Pretoria, South Africa: Department of Agriculture, Forestry and Fisheries, South Africa (DAFF); 1996. Available from: http://www.daff.gov.za/.
62. Dweba CC, Zishiri OT, El Zowalaty ME. Methicillin-resistant Staphylococcus aureus: livestock-associated, antimicrobial, and heavy metal resistance. Infect Drug Resist. 2018;11:2497.
63. Igbinosa IH. Prevalence and detection of antibiotic-resistant determinant in Salmonella isolated from food-producing animals. Trop Anim Health Prod. 2015;47(1):37–43. doi:10.1007/s11250-014-0680-8
64. Madoroba E, Kapeta D, Gelaw AK. Salmonella contamination, serovars and antimicrobial resistance profiles of cattle slaughtered in South Africa. Onderstepoort J Vet Res. 2016;83(1):1–8. doi:10.4102/ojvr.v83i1.1109
65. Mathole M, Muchadeyi F, Mdladla K, Malatji D, Dzomba E, Madoroba E. Presence, distribution, serotypes and antimicrobial resistance profiles of Salmonella among pigs, chickens and goats in South Africa. Food Control. 2017;72:219–224. doi:10.1016/j.foodcont.2016.05.006
66. Kuang D, Xu X, Meng J, et al. Antimicrobial susceptibility, virulence gene profiles and molecular subtypes of Salmonella Newport isolated from humans and other sources. Infect Genet Evol. 2015;36:294–299. doi:10.1016/j.meegid.2015.10.003
67. Iwu CJ, Iweriebor BC, Obi LC, Basson AK, Okoh AI. Multidrug-resistant Salmonella isolates from swine in the Eastern Cape Province, South Africa. J Food Prot. 2016;79(7):1234–1239. doi:10.4315/0362-028X.JFP-15-224
68. Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect Drug Resist. 2015;8:49. doi:10.2147/IDR
69. Phillips I, Casewell M, Cox T, et al. Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J Antimicrob Chemother. 2004;53(1):28–52. doi:10.1093/jac/dkg483
70. Ahmed AM, Shimabukuro H, Shimamoto T. Isolation and molecular characterization of multidrug‐resistant strains of Escherichia coli and Salmonella from retail chicken meat in Japan. J Food Sci. 2009;74(7):M405–M410. doi:10.1111/j.1750-3841.2009.01291.x
71. VT Nair D, Venkitanarayanan K, Kollanoor Johny A. Antibiotic-Resistant Salmonella in the Food Supply and the Potential Role of Antibiotic Alternatives for Control. Foods. 2018;7(10):167. doi:10.3390/foods7100167
72. Møller TS, Overgaard M, Nielsen SS, et al. Relation between tetR and tetA expression in tetracycline resistant Escherichia coli. BMC Microbiol. 2016;16(1):39. doi:10.1186/s12866-016-0649-z
73. Sørensen JT, Edwards S, Noordhuizen J, Gunnarsson S. Animal production systems in the industrialised world. Rev Sci Tech. 2006;25(2):493–503.
74. Eguale T, Birungi J, Asrat D, et al. Genetic markers associated with resistance to beta-lactam and quinolone antimicrobials in non-typhoidal Salmonella isolates from humans and animals in central Ethiopia. Antimicrob Resist Infect Control. 2017;6(1):13. doi:10.1186/s13756-017-0171-6
75. Giuriatti J, Stefani LM, Brisola MC, Crecencio RB, Bitner DS, Faria GA. Salmonella Heidelberg: genetic profile of its antimicrobial resistance related to extended spectrum β-lactamases (ESBLs). Microb Pathog. 2017;109:195–199. doi:10.1016/j.micpath.2017.05.040
76. Meissner H, Scholtz M, Palmer A. Sustainability of the South African Livestock Sector towards 2050 Part 1: worth and impact of the sector. S Afr J Anim Sci. 2013;43(3):282–297. doi:10.4314/sajas.v43i3.5
77. Kiiru J, Butaye P, Goddeeris BM, Kariuki S. Analysis for prevalence and physical linkages amongst integrons, ISE cp 1, IS CR 1, Tn 21 and Tn 7 encountered in Escherichia coli strains from hospitalized and non-hospitalized patients in Kenya during a 19-year period (1992–2011). BMC Microbiol. 2013;13(1):109. doi:10.1186/1471-2180-13-109
78. Argüello H, Guerra B, Rodríguez I, Rubio P, Carvajal A. Characterization of antimicrobial resistance determinants and class 1 and class 2 integrons in Salmonella enterica spp., multidrug-resistant isolates from pigs. Genes. 2018;9(5):256. doi:10.3390/genes9050256
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
