Back to Journals » Infection and Drug Resistance » Volume 16
Molecular Characterization of Extensively Drug Resistant Salmonella Enterica Serovar Typhi Clinical Isolates from Lahore, Pakistan
Authors Jabeen K, Saleem S, Jahan S , Nizamudin S, Arshad F , Huma ZE, Raza SM, Mehmood M, Roman M, Haq FU
Received 10 February 2023
Accepted for publication 11 April 2023
Published 12 May 2023 Volume 2023:16 Pages 2987—3001
DOI https://doi.org/10.2147/IDR.S406253
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Kokab Jabeen,1 Sidrah Saleem,1 Shah Jahan,1 Summiya Nizamudin,2 Faiqa Arshad,1 Zill-e Huma,1 Syed Mohsin Raza,3 Maria Mehmood,1 Muhammad Roman,1 Faiz Ul Haq1
1Department of Microbiology, University of Health Sciences, Lahore, Pakistan; 2Department of Microbiology, Shaukat Khanum Memorial Cancer Hospital & Research Centre, Lahore, Pakistan; 3Department of Allied Health Sciences, University of Health Sciences, Lahore, Pakistan
Correspondence: Kokab Jabeen, Tel +92 3128878893, Email [email protected]
Background: The emergence of extensively drug-resistant (XDR) typhoid in Pakistan has endangered the treatment options available to manage this infection. Third generation cephalosporin were the empiric choice to treat typhoid fever in Pakistan, but acquisition of ESBLs have knocked them out of the arsenal. The current empiric choice is azithromycin which is vulnerable to resistance too. This study aimed to assess the burden of XDR typhoid and the frequency of resistance determinants in blood culture samples collected from different hospitals in Lahore, Pakistan.
Methods: A total of 835 blood cultures were collected from different tertiary care hospitals in Lahore during January 2019 to December 2021. Among 835 blood cultures, 389 Salmonella Typhi were identified, and 150 were XDR S. Typhi (resistant to all recommended antibiotics). Antibiotics resistance genes of the first-line drugs (blaTEM-1, catA1, sul 1, and dhfR7) and second line drugs (gyrB, gyrA, qnrS, ParC and ParE) were investigated among XDR S. Typhi. There were different CTX-M genes isolated using the specific primers, blaCTX-M-U, blaCTX-M-1, blaCTX-M-15, blaCTX-M-2, blaCTX-M-8 and blaCTX-M-9.
Results: Antibiotic resistant genes of the first-line drugs were isolated with different frequency, blaTEM-1 (72.6%), catA1 (86.6%), sul 1 (70%), and dhfR7 (56%). Antibiotics resistance genes of second-line drugs were isolated as: gyrB (60%), gyrA (49.3%), qnrS (32.6%), parC (44%) and parE (28%). Among CTX-M genes, blaCTX-M-U (63.3%) was the most frequent followed by blaCTX-M-15 (39.3%) and blaCTX-M-1 (26%).
Conclusion: Our study concluded that XDR isolates circulating in Pakistan have acquired first-line and second-line antibiotic resistant genes quite successfully along with CTX-M genes (ESBLs) rendering them resistant to the third generation cephalosporins as well. Emergence of azithromycin resistance in XDR S. Typhi which is currently used as an empiric treatment option is worrisome and needs to be monitored carefully in endemic countries like Pakistan.
Keywords: Salmonella, antimicrobial resistance, plasmids, XDR typhoid
Introduction
Typhoid fever is a prominent public health concern in low and middle-income countries, including Pakistan. Approximately 11 to 21 million cases of typhoid were reported worldwide, with 128,000 to 161,000 deaths.1 Residents of the Punjab and Sindh provinces are the most vulnerable to contracting typhoid among the 16 Asian nations where the disease is endemic.2 According to a study reported in 2018, Pakistani citizens were estimated to have the highest infection rate of Salmonella Typhi among all other South East Asian countries. The reported cases per 100,000 population were almost 493.5.3 In 2016, the southeastern province of Pakistan (Sindh) reported the first large-scale outbreak.4 Similar reports from Punjab Province5–7 and international travelers were soon being published.8–10 In specific regions of Pakistan where prevailing conditions exist, the prevalence of XDR typhoid has increased from 7/100,000 to 15/100,000.2,11 Since September, 2020, 2883 cases have been reported in Pakistan and all of them were extensively resistant isolates.12 According to an estimate by WHO, almost 10,365 cases of XDR S. Typhi have been identified by 2019.13
Prior to this outbreak, there were sporadic cases of extended spectrum beta-lactamase producing S. Typhi reported from different places. Africa, Netherlands, France, Senegal, Latin America, Asia, and Europe reported cases of ceftriaxone-resistant isolates of S. Typhi.14,15 Bangladesh, India, and Pakistan have reported isolated cases of S. Typhi, resistant to third-generation cephalosporins.16 The emergence of an ESBL in S. Typhi poses a new contest and was a major cause for unease, particularly in developing countries.17,18 Prior to the development of this clone, ceftriaxone resistance was not widespread in S. Typhi, although it had been reported in other serovars such as S. Typhimurium, S. Stanley, and others.
From 2009 to 2011, laboratory monitoring data from Karachi, Pakistan, revealed an increase in multidrug-resistant S. Typhi and a very low incidence of occasional third-generation cephalosporin resistance ie 0.08%.19 Laboratory surveillance is the most effective method for detecting a rise in antibiotic resistance, so the fact that this percentage is so low is concerning to the medical community. Following the publication of a report from Sindh province, similar cases were reported from Punjab province. The most prevalent reported variant was blaCTXM-15.5,6 In addition to Pakistan, neighboring countries including India, Bangladesh, Nepal, and Sri Lanka have reported similar cases of S. Typhi strains producing ESBL. In a recent study from India, 25% of S. Typhi isolates showed the production of ESBLs, and CTX types 2 and 9 were the most prevalent.19–21
The emergence of XDR S. Typhi has created an enormous therapeutic challenge for physicians around the globe. The only oral option available to treat this superbug is azithromycin. Fearing its misuse in the current COVID-19 pandemic, acquiring a single resistance can change the treatment paradigm of typhoid fever from outpatient to hospitalization of every patient.22
Though azithromycin resistance is uncommon in S. Typhi, sporadic resistance to Salmonella species has been reported from all over the world. There are reports of clinical treatment failures, and antimicrobial resistance in the laboratory from different parts of the world.23 In a recent study from Karachi, Pakistan, where an XDR S. Typhi outbreak is happening, an alarming rate of azithromycin resistance (33%) was reported.24 S. Typhi and ParaTyphi A isolates were found to be resistant to azithromycin (MIC range: 32–64 µg/mL) in a Bangladeshi study. Using whole-genome sequencing data, it was impossible to identify the molecular basis of azithromycin resistance.25
Typhoid conjugate vaccines have been incorporated into routine immunization programs in five countries (Liberia, Nepal, Pakistan, Samoa, and Zimbabwe) with estimated high typhoid fever incidence (100 cases per 100,000 population per year), high antimicrobial resistance prevalence, or recent outbreaks since 2018. WHO advises that countries with endemic typhoid fever establish health facility-based surveillance with laboratory confirmation to assess the effectiveness of vaccines, monitor patterns of antimicrobial resistance, and determine the disease burden.26 The emergence and spread of Salmonella enterica subsp. enterica serovar Typhi lineages, which are resistant to many essential antimicrobial agents for therapy, complicate the treatment of typhoid fever. S. Typhi lineages with varying degrees of resistance to first- and second-line typhoid antimicrobial agents, including ampicillin (AMP), chloramphenicol, trimethoprim-sulfamethoxazole (SXT), ciprofloxacin (CIP), third-generation cephalosporins, and azithromycin, have emerged globally as a result of selection pressure from antibiotic use in regions where typhoid fever is endemic.27,28 In Enterobacteriaceae, carbapenem resistance is uncommon and likely first observed in S. Typhi from Faisalabad, Pakistan. The 2018 S. Typhi isolates contained the H58 haplotype, and the PCR-restriction fragment length polymorphism method determined that 16.7% of the H58 strains belonged to lineage I, 19.4% to lineage II, and the remaining 63.9% to the node. Effective epidemiological studies are required to address the regional variation in the antimicrobial resistance trend.29
Previous studies on Salmonella from Lahore, Pakistan only focus on epidemiology, antimicrobial susceptibility testing (AST), detection of ESBL and these are usually from single centers with a small number of samples6,22,30,31 Here we extended our study with collection of samples from all the major tertiary care hospitals in Lahore. In addition to this we used standard broth microdilution method and E-strips for AST and all the 150 XDR S. Typhi isolates were subjected to molecular analysis to get insight into the underlying molecular mechanism of resistance. Lahore lacks laboratory data regarding the current sensitivity pattern of azithromycin against typhoidal Salmonella. Laboratory surveillance is a crucial tool for predicting the emergence of resistance to any antibiotic. Consequently, it is urgent to investigate the current susceptibility profile of typhoidal salmonellae, particularly XDR S. Typhi, which is currently treated with azithromycin alone.
Materials and Methods
Study Design and Setting
A cross-sectional descriptive study was conducted at the Microbiology and the Immunology departments, University of Health Sciences, Lahore, Pakistan from January 2019 to December 2021.
Sample Collection, Culturing and Identification of Bacteria
In this study eight hundred and thirty five (n=835) blood cultures were collected as part of hospital routine procedures from different tertiary care hospitals in Lahore and BacT/ALERT3D microbial Detection System (BioMérieux, France) was used. Bottles identified as positive by the BacT/ALERT3D system were subcultured on MacConkey and blood agar plates. Inoculated plates were incubated aerobically for 18–24 h in inverted position at 37°C. If after 24 h no growth was observed visually the plates were incubated again for an additional 24 h. The standard laboratory operating procedures for identifying the organism included colonial morphology, gram staining, serological and biochemical tests using the AP1–20E identification system (BioMérieux, France). Salmonella O, H, and Vi antigen (BD Difco, USA) was used to perform serological identification of S. Typhi according to the manufacturer’s instructions.
Antimicrobial Susceptibility Testing
Using commercially available antimicrobial discs, the antimicrobial susceptibility pattern of S. Typhi was performed using the Kirby–Bauer disc diffusion method using Mueller–Hinton agar (Oxoid, UK), in accordance with the Clinical and Laboratory Standard Institute 2021 (CLSI) guidelines. Antimicrobial disks with standard antibiotic content were used (Oxoid, USA). For all antimicrobial susceptibility tests, Escherichia coli (ATCCR 25922) and Pseudomonas aeruginosa (ATCCR 27853) was used as the control strain.
Determination of Minimum Inhibitory Concentrations (MICs) of Recommended Antibiotics
MICs of antibiotics against XDR isolates were determined using the automated VITEK 2 compact system (BioMe´rieux, France) and interpretation was carried out as per the clinical laboratory standards institute (CLSI) 2021 guidelines (https://clsi.org). S. Typhi that had high MICs against ceftriaxone and cefotaxime (>4 µg/mL) were considered as XDR. Staphylococcus aureus (ATCC 29213) was used for quality control of E-strip method for azithromycin as per recommendation.
MICs of azithromycin were determined by E-strips according to manufacturer’s instructions (Liofilchem, Italy). Bacterial suspension was adjusted according to 0.5 McFarland’s standard. Isolates were inoculated on the surface of Mueller–Hinton agar plate using a sterile cotton swab. E-strips were applied to the surface of agar by using sterile forceps. After placing E-strips, plates were incubated at 37°C aerobically for 18–24 h. MIC results were interpreted according to CLSI 2021 guidelines. For azithromycin MIC >32 µg/mL was taken as resistant.
Molecular Confirmation of H58 Haplotype, S. Typhi and XDR S. Typhi
Isolates phenotypically identified as S. Typhi were molecularly confirmed by PCR amplification of the target genes. Molecular identification of S. Typhi, XDR S. Typhi and S. Typhi H58 haplotype was carried out using specific primers.32
Molecular Detection of Antibiotic Resistant Genes Including Azithromycin Resistant Genes
Molecular identification of antibiotic resistant genes of first-line drugs was performed using the specific primers: blaTem-1, catA1, sul1, and dhfR7 were amplified by PCR. Antibiotic resistant genes of second-line drugs were isolated using the specific primers for gyrB, gyrA, qnrS, ParC and ParE. Different CTX-M genes were isolated using the specific primers, blaCTX-M-U,blaCTX-M-1,blaCTX-M-15,blaCTX-M-2,blaCTX-M-8 and blaCTX-M-9.5,6,33 Molecular identification of azithromycin resistant genes (mefA, msrA, macA, mphA, mphB, ermA, ermB, ermC, ereA, ereB, and acrB) was carried out using specific primers in three azithromycin resistant isolates.34,35 Sequences of the primers are given in the Table S1–S7.
The reaction mixture of 25 µL for a single reaction included the following: master mix 12.5 µL (1X) (DreamTaq DNA polymerase, 2X DreamTaq green buffer, dATP, dCTP, dGTP and dTTP, 0.4 mM each, and 4 mM MgCl2), forward and reverse primers 0.5 µL (0.2 µM) each, DNA 2 µL (50 ng), dH2O (nuclease-free) 9.5 µL. Negative control was made by adding all mixture content excluding DNA. Positive control was made by adding extracted DNA of all XDR isolates. After optimization, amplification of the DNA fragments was performed in a thermal cycler (Bio-Rad PCR system T100TM 621BR 35960).
The cycling conditions for PCR were 1 cycle of pre-denaturation at 94°C for 5 min, 35 cycles each of denaturation at 94°C for 1 min, annealing at different optimized temperatures for different genes and extension at 72°C for 1 min. This was followed by final extension of one cycle at 72°C for 5 min and final holding at 4°C. The PCR products were analyzed using gel electrophoresis (Thermofisher Scientific).
Statistical Analysis
Statistical analysis of the data was performed by using Statistical Package for Social Sciences (SPSS) software version 26. The presentation of categorical data is in the form of frequencies and percentages. Chi-squared test was applied to conduct comparative analysis among different sets. A P-value of ≤0.5 was taken as statistically significant. The heatmap was created in R package by the basic heatmap function. A distance matrix was produced using UPGMA clustering (http://genomes.urv.cat/UPGMA), and a picture was envisioned using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). On Adobe Illustrator CS5 version 15.0.0, trees and graphs were labeled or colored differently to make visualization of variants and their characteristics possible (Adobe Systems, Inc., San Jose, CA, USA). CorrPlot R package and the basic hclust R function were used to construct correlation between antibiotics and resistant genes.
Results
Distribution of S. Typhi and XDR S. Typhi in Blood Cultures
Among 835 blood cultures, 389 (47%) were identified as S. Typhi. During 2019–20 isolation frequency was 34%, while it increased by 57% during2020–21. A rising trend was seen in isolation frequency of S. Typhi in blood cultures (P<0.05) (Table 1). Out of these 389 S. Typhi, 150 (40%) were identified as extensively drug resistant (XDR), 126 (32%) as multidrug resistant (MDR), and only three (2%) as azithromycin resistant (AZM resistant), on the basis of their antimicrobial susceptibility testing (Table 1). Out of 150 XDR isolates, 70 (46%) were collected in year 2019–20, whereas 80 (54%) were collected during 2020–21 (Table 1).
 |
Table 1 Distribution of XDR S. Typhi in Blood Cultures (n=835) |
Antimicrobial Sensitivity Testing and Phenotypic Confirmatory Testing of XDR S. Typhi
All XDR isolates 150 (100%) were resistant to ampicillin, ciprofloxacin, ceftriaxone, cefixime and cefepime according to CLSI 2021 guidelines, as shown in Table 2. There were only four (2.7%) isolates sensitive to chloramphenicol. Another first-line drug co-trimoxazole showed resistance in 146 (97.3%) isolates. Most effective antibiotics were carbapenems ie imipenem and meropenem with sensitivity of 100%. Out of all isolates, only three (2%) were resistant to azithromycin with MIC of ˃32 µg/mL. Piperacillin-tazobactam was sensitive in all the isolates except three (2%) were resistant. Mean MIC of antibiotics against XDR S. Typhi were shown in Figure 1. Ampicillin and chloramphenicol showed ≥32.00 µg/mL against XDR S. Typhi which were resistant according to CLSI 2021. The MIC of ciprofloxacin was 03.22±0.07 µg/mL and ceftriaxone were 25.39±01.16 µg/mL.
 |
Table 2 MIC Distribution of Various Antibiotics in XDR S. Typhi |
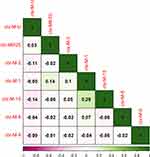
Heatmap in R
A rearranged heatmap was created to represent the resistance phenotypes of every isolate according to the colored code given and shown in the Figure 2 key (red = resistant and blue = sensitive). Four clusters of interest representing different resistance patterns were visible representing few sensitive isolates to chloramphenicol and trimethoprim-sulfamethoxazole in the bottom portion where all other isolates were resistant to these antibiotics. The other two clusters in top portion represent resistance where all other isolates were sensitive to these plotted drugs except TZP and AZI.
Molecular Confirmation of S. Typhi, XDR S. Typhi and H58 Haplotype
All 150 (100%) isolates harbored target genes confirming their origin from H58 haplotype. The presence of specific genes also confirmed the phenotypically identified S. Typhi and XDR S. Typhi.
Molecular Basis of Resistance to First-line Antibiotics
Among first-line antibiotics resistant genes, catA1 was the most prevalent 130 (86.6%) followed by blaTEM1 109 (72.6%), and sul1 105 (70%). The gene, dhfR7 was only detected among 84 (56%) XDR isolates of S. Typhi as shown in Table 3. We believed that XDR S. Typhi has adopted first-line antibiotic-resistant genes present in MDR S. Typhi.
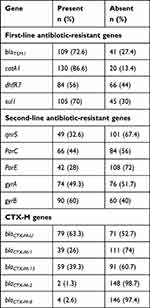 |
Table 3 Frequency of First-line Antibiotics, Fluoroquinolone Resistant, and CTX-M Genes |
Molecular Basis of Resistance to Second-line (Fluoroquinolone) Antibiotics
Fluoroquinolone resistance in S. Typhi has been attributed to the one or more point mutations in quinolone resistant determining region. The most frequently reported AMR genes are gyrB 90 (60%), gyrA 74 (49.3%), parC 66 (44%), and parE 42 (28%). In addition acquisition of IncY plasmid related qnrS has been implicated in resistance against fluoroquinolones especially in 49 (32.6%) XDR isolates (Table 3).
Molecular Basis of Resistance to Third-generation Cephalosporins
Third-generation cephalosporin resistance has been attributed to an IncY plasmid carrying blaCTX-M-15 gene. But in our study we screened for the presence of additional CTX-M variants. CTX-M gene blaCTX-M-U was detected among 79 (63.3%) S. Typhi isolates, while blaCTX-M-1 was present in 39 (26%) isolates as shown in Table 3.
Genotype-phenotype Correlation
A distance matrix was produced using UPGMA clustering (http://genomes.urv.cat/UPGMA), and a tree was visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). On Adobe Illustrator CS5 version 15.0.0, trees and graphs were labeled or colored differently to make visualization of variants and their characteristics possible (Adobe Systems, Inc., San Jose, CA, USA).
UPGMA clustering lead to the formation of different clusters, according to genotype and phenotypes of the isolates. Trees illustrated relationship between diverse isolates according to their antibiotic susceptibility phenotypes (Figure 3). Circular trees were created by the unweighted pair group with arithmetic mean method (UPGMA). Figures were abridged as 1/0 values representative of 10 resistant phenotypes and seven resistant genotypes (Figure 3).
 |
Figure 3 Circular trees created by the UPGMA (unweighted pair group method with arithmetic mean) method (http://genomes.urv.cat/UPGMA). This figure is showing UPGMA clustering lead to the formation of different clusters, according to genotype of the isolates. Trees illustrate the relationship between diverse isolates according to their beta-lactamase genotypes. Branch labels are mentioned at the bottom as ±, while tip labels are color-coded according to presence of CTXM genes. |
Tree tips (isolates IDs) are coded with different colors conferring to their blaCTX-M genotype (indicated in the figure key). Twigs of the genotype tree (Figure 4) are colored and then labeled according to their genotype identity (+/−patterns); while three major clades (Figure 4) are colored according to their resistance pattern in the phenotype tree.
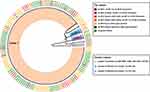 |
Figure 4 Circular trees created by the UPGMA (unweighted pair group method with arithmetic mean) method (http://genomes.urv.cat/UPGMA). This figure is showing UPGMA clustering lead to the formation of different clusters, according to antimicrobial susceptibility of isolates and CTX-M genotype. Cluster labels are mentioned at the bottom as different colors, while tip labels are color-coded according to presence of CTXM genes in these clusters. |
Cluster 1 was the most prominent; these isolates were sensitive to IMP (imipenem), MER (meropenem), AZM (azithromycin), CEF-AVI (ceftazidime-avibactam) and TZP (piperacillin-tazobactam). Cluster 2 was sensitive to cluster 1 and CHL (chloramphenicol). Isolate IDs in this cluster were, UHS 070, UHS 079, UHS 102 and UHS 138. All of these isolates except UHS138 had one or more CTX-M genes present in them and phenotypically were XDR isolates. Cluster 3 comprising of isolate IDs, UHS 074, UHS 088, UHS 110 and UHS 139 were sensitive to cluster 1 and COT (co-trimoxazole). All of these isolates were phenotypically XDR isolates with resistance to all first-line, second-line and third-line drugs except co-trimoxazole.
CorrPlot Correlation Between First-line, Second-line Antibiotic-resistant and CTX-M Genes
A false colored disproportionate relationship medium representing the correlations among each antibiotic resistant gene pairs according to their presence in all 150 isolates was made using the basic hclust R function and the CorrPlot R package (Figures 5 and Figure 6). For each pair of genes, Spearman’s rank correlation coefficients (rs) are shown.
The genes are clustered hierarchically, and the grouping is signified by the tree. Tree branches represent 1-rs. The Spearman's correlation coefficient (rs) was 1 for same pair of genes. The significant value of Spearman's correlation coefficient (rs) was ≥0.5. This means that genes having value of ≥0.5 have a positive correlation. The analysis indicated that catA1and gyrB, parE and qnrS, gyrA and Sul1 and dhfR7 and gyrA were the most correlated genes. All of them had value of Spearman's correlation coefficient (rs) =1. However, there was no significant correlation between CTX-M genes as all of them had value of Spearman's correlation coefficient (rs) <0.5 (Figure 6).
Discussion
XDR S. Typhi outbreak started from Sindh province in 2016, has taken over the Punjab province as well. In this study antibiotic resistance genes were investigated located in the IncHI1 (catA1, blatem1, dhfR7, and sul1), QDRD (gyrB, gyrA, qnrS, ParC and ParE) and IncY (qnrS and blaCTX-M-15) regions, which contribute to resistance to first and second-line antibiotics, respectively, in order to understand the molecular basis of the antibiotic resistance mechanisms among these XDR S. Typhi.
Our MIC results and other findings correlated with the molecular determinants of MDR and XDR phenotypes among the recently reported XDR S. Typhi isolates from typhoid patients, except for azithromycin.4,5 With limited treatment options, emerging azithromycin resistance would create a further problem. In the present study frequency of azithromycin resistant XDR isolates was found to be 2%. During 2019–20 it was 0.67%, while during 2020–21 it was 1.33%. This showed a significant increasing trend in azithromycin-resistant isolates by 140% from the years 2019–20 to 2020–2021. An explanation of this finding could be the unwarranted and unjudicial use of this antibiotic for the treatment of the COVID-19 pandemic in Pakistan where there is over- the-counter availability of multiple antibiotics.
A report from Lahore, Pakistan by Ahmad et al reported 7.3% azithromycin resistance in XDR isolates which was quite higher than the present study.11 Hussain et al from Karachi (2019), reported 3% azithromycin resistance in XDR S. Typhi.36 A similar report stated 2% azithromycin resistant XDR isolates from CMH, Lahore.37 Another study from Agha khan hospital, Karachi reported 4.5% azithromycin treatment failure cases in XDR S. Typhi.38 An alarming study from Karachi,24 reported 32.8% resistance to azithromycin in XDR which was very high compared to the present study.
Typing antibiotic-resistant bacterial strains can help identify clonal spread. The S. Typhi H58 haplotype is globally associated with MDR. A previous study confirmed haplogroup diffusion with 47.4% of H58 S. Typhi.29 In our study all 150 (100%) isolates were of H58 haplotype, also called 4.3.1 genotype confirming that there was smooth transition from MDR to XDR in this most prevalent haplotype. This haplotype is dominant in South Asia as well as globally as reported by Jamilah et al.39 This haplotype has been reported to be most prevalent in the South Asia, Africa, Indonesia, UK, Zimbabwe, and Australia.40 So this could help in global spread of this XDR clone.
Molecular determinants of first-line and second-line antibiotic resistance in XDR isolates were isolated with variable frequency. For ampicillin resistance blaTem-1 was found in 72.6% isolates and for chloramphenicol resistance, catA1 gene was detected in 86.6% isolates. For trimethoprim-sulfamethoxazole, dhfR7 and sul1 were detected in 56% and 70% isolates, respectively. A recent study from Lahore, Pakistan reported 42 (51%) isolates were blaTem-1 positive.6 Another study reported that all 18 XDR isolates harbored blaTem-1.5 Regarding fluoroquinolone resistance genes, gyrB was the most frequent gene present in 60% isolates, gyrA, qnrS, ParC and ParE were present in 49.3%, 32.6%, 44% and 28% isolates, respectively. Recently, Chen et al used whole genome sequencing (WGS) for the detection of antimicrobial resistance genes in non-typhoidal Salmonella, who detected of 50 genes encoding resistance to 11 different categories in addition to four mutations affecting the gyrA gene.28
An outbreak of XDR S. Typhi was recently reported in China, with five XDR S. Typhi isolates recovered from water sources. These five XDR S. Typhi isolates all carried the same IncY plasmid recovered by WGS, which contained a Type 4 secretion system gene cluster and six resistance elements, including sul2, blaTem-1B, blaCTX-M-15, and the qnrS fluoroquinolone resistant gene.41
A report published in 2019 exploring genetic profiling of antimicrobial resistant genes in S. Typhi isolates from Bangladesh stated that blaTem-1 was present in ampicillin sensitive isolates and was absent in some resistant isolates, similar to our study.42 Its absence could suggest alternate mechanisms of resistance such as presence of blaTem-1B.41 A study established that the presence of beta-lactam genes was associated with ampicillin resistance in 15.79% of the isolated strains tested by WGS, blaTem-1B, the most common beta-lactam resistant gene, was found in 19 strains, blaTem-1 in 16 strains, and blaTem-116 in one strain.43 A recent study from Lahore, reported that 121 (67%) of the Salmonella isolates were MDR, and 62 (34%) were XDR. The ampC gene, which has a 47% prevalence, was the most common resistance gene, followed by the gyrA, catA1, tet(A), aac (3)-la, qnrS, blaNDM-1, and blaCTX-M-15 genes in isolates with 45, 40, 21, 6, 18, 3, 11, 6, and 2%, respectively.44 This study did not report presence of blaTEM-1 or any other variants in their XDR isolates.
Mobile genetic elements, which transfer resistance and virulence genes between bacteria, are major contributors to the rapid spread of antimicrobial resistance.28 Regarding resistance determinants of third-generation cephalosporin, in our study we reported different types of CTX-M for the very first time. Many studies have reported high prevalence of blaCTX-M-15 in XDR isolates5,41,44–46 but in our study other types were also identified. In our study blaCTX-M-U was the most frequent gene present in 63.3% XDR isolates followed by blaCTX-M-15 (39.3%). blaCTX-M-1, blaCTX-M-2, blaCTX-M-8 and blaCTX-M-9 present in 26%, 1.3%, 2.6%, 0.6% respectively. Previously the beta-lactamase gene was reported including blaSHV-12 from Norway and the Netherlands, blaCTX-M-U from Bangladesh, and blaCTX-M-15 from UAE.14,47–49 This could possibly explain findings of our study and presence of CTX-M variants and this highlights the importance of regional and geographical variation in genotyping. Comparative genomic analysis and thorough plasmid characterization show that in India (4.3.1 lineage II), ceftriaxone resistance is caused by the short-term persistence of resistance plasmids like IncX3 harboring blaSHV-12 or IncN having blaTEM-1B and blaDHA-1, whereas in Pakistan (4.3.1 lineage I), the XDR trait is associated with the blaCTX-M-15 gene on IncY plasmid.50
Azithromycin is the only oral treatment option available to treat XDR typhoid. Other treatment options are injectable carbapenems (imipenem and meropenem). Our XDR isolates were 100% susceptible to carbapenems, but 2% were resistant to azithromycin with MIC value >32 µg/mL which is quite alarming. Reports of carbapenem resistance are quite rare in this part of the world as well as globally. In a recently reported study from Punjab, Pakistan, 48% isolates were resistant to carbapenems.51 Global data from various countries reported 2.5% resistance to carbapenems in S. Typhi isolates between 1972 and 2018.52 The results of this study support the idea that host variation and transmission procedures may differ in patients belonging to different geographical locations. These findings also suggest that, over time, nearly all locally circulating S. Typhi would likely adopt molecular determinants for azithromycin and carbapenem resistance and this could be a global phenomenon in near future.
Our current study indicates that our XDR isolates do not carry any molecular determinants that have been attributed to the azithromycin resistance in typhoidal Salmonellae. Regarding molecular determinants of azithromycin resistance we tried to detect all possible genes, but were not able to identify any molecular determinants of resistance. In this study no azithromycin resistant XDR isolates harbored, acrB, mphA, mphB, ermA, ermB, ermC, ereA, ereB, mefA, msrA and macA genes. A recent report from India reported 0.75% azithromycin resistance in MDR S. Typhi with presence of R717Q mutation on the acrB efflux pump by PCR.43
A recent study from Bangladesh reported azithromycin resistance among MDR isolates, conferred by acrB in 54 isolates across eight different genotypes that seemed to be spontaneous emergence of azithromycin resistance in Bangladeshi population highlighting the importance of population diversity and epidemiological factors.46 The apparent mechanism of higher MIC against azithromycin was a mutation in the gene encoding acrB (R717Q).53 Presence of R717Q mutation in the acrB gene was thought to be spontaneous mutant rather than a clone of resistant isolates spreading.54
Macrolide resistant genes have been identified as cause of azithromycin resistance in enteric bacteria like E. coli. The plasmid borne mphA gene found in azithromycin resistant S. sonnei, was the commonest macrolide resistance gene found in E. coli. The gene was mostly found in isolates from patients who had received antimicrobial drugs or had been hospitalized, particularly in ESBL producers.34 This study highlighted the chances of its spread to Salmonella species especially S. Typhi as this is a target of azithromycin as well. But to our knowledge none of the study has reported these genes in azithromycin resistance isolates to date including our study.
In some S. Typhi and S. ParaTyphi A clinical isolates, a point mutation or mutations on the antibiotic binding subunit (AcrB) of the tripartite efflux pumps (AcrAB-TolC), such as R717Q or R717L, has recently been linked to azithromycin resistance.55 Point mutations on the promoter of macA of the tripartite efflux pump (macAB-TolC) have been linked to azithromycin resistance in Neisseria gonorrhoeae, a gram-negative bacterial pathogen similar to S. Typhi.56
S. Typhi resistance to macrolides is a serious global health concern that requires close monitoring for local and global spread. We were not able to screen molecular determinants of azithromycin resistance but development of phenotypic resistance can change the paradigm of XDR typhoid management in endemic countries like Pakistan.
The current study has several limitations. First, we did not look into Salmonella resistance mechanisms using whole-genome sequencing. Because this is a hospital-based study that is only limited to S. Typhi, we recommend that nontyphoidal Salmonella be studied for the molecular basis of drug resistance. Although outside the scope of the current study, additional animal studies would have been useful in predicting resistant mechanisms and possible antibiotic resistant links.
Conclusions
It is clearly evident that MDR, XDR S. Typhi along with the emergence of azithromycin resistance is present in the Punjab province. Almost all XDR S. Typhi have acquired resistance to the first-line and second-line antibiotics and to the third generation cephalosporins. The potential azithromycin resistance in XDR Salmonella Typhi is imminent and may be attributed to its extensive use in the COVID-19 pandemic. Sensitivity patterns should be reported with MICs values, and if possible higher values should be screened for the presence of any potential resistance genes. Antibiotic stewardship should be strictly implemented. Some of the methods used in the current study to detect key molecular determinants of the antibiotic resistance in S. Typhi could be developed as a surveillance and point-of-care testing strategy. Future surveillance could include additional molecular traits predicted to be associated with resistance to macrolides as described in this study.
Data Sharing Statement
The datasets used and/or analyzed during the current study are not publically available because study was completely anonymous, any demographic data or identifiable information was not obtained therefore informed consent was not required this type of study according to the local legislation. This study was conducted using the bacterial strains which were isolated for diagnostic and treatment purpose. However it is available from the corresponding author on reasonable request.
Ethical Approval
The study was ethically approved by the “Ethical Review Board” under reference number UHS/REG-19/ERC/398 from University of Health Sciences, Lahore, Pakistan.
Acknowledgments
We would like to acknowledge all the technical staff of the Microbiology Department, University of Health Sciences, Lahore and all the tertiary care hospitals from which we collected samples, for their help, and technical assistance.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J Med Soc. 2017;2017:1–7.
2. Rasheed MK, Hasan SS, Ahmed SI, Ahmed SI. Extensively drug-resistant typhoid fever in Pakistan. Lancet Infect Dis. 2019;19(3):242–243. doi:10.1016/S1473-3099(19)30051-9
3. Fatima M, Kumar S, Hussain M, et al. Morbidity and mortality associated with typhoid fever among hospitalized patients in Hyderabad District, Pakistan, 2017–2018: retrospective record review. MIR Public Health Surveill. 2021;7(5):e27268. doi:10.2196/27268
4. Klemm EJ, Shakoor S, Page AJ, et al. Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. MBio. 2018;9(1):e00105–00118. doi:10.1128/mBio.00105-18
5. Kim C, Latif I, Neupane DP, et al. The molecular basis of extensively drug-resistant Salmonella Typhi isolates from pediatric septicemia patients. PLoS One. 2021;16(9):e0257744. doi:10.1371/journal.pone.0257744
6. Saeed M, Rasool MH, Rasheed F, et al. Extended-spectrum beta-lactamases producing extensively drug-resistant Salmonella Typhi in Punjab, Pakistan. J Infect Dev Ctries. 2020;14(02):169–176. doi:10.3855/jidc.12049
7. Nizamuddin S, Ching C, Kamal R, Zaman MH, Sultan F. Continued outbreak of ceftriaxone-resistant Salmonella enterica serotype typhi across Pakistan and assessment of knowledge and practices among healthcare workers. Am J Trop Med Hyg. 2021;104(4):1265–1270. doi:10.4269/ajtmh.20-0783
8. Watkins LKF, Winstead A, Appiah GD, et al. Update on extensively drug-resistant Salmonella serotype Typhi infections among travelers to or from Pakistan and report of ceftriaxone-resistant Salmonella serotype Typhi infections among travelers to Iraq—United States, 2018–2019. Morb Mortal Weekly Rep. 2020;69(20):618. doi:10.15585/mmwr.mm6920a2
9. Chirico C, Tomasoni LR, Corbellini S, et al. The first Italian case of XDR Salmonella Typhi in a traveler returning from Pakistan, 2019: an alert for increased surveillance also in European countries? Travel Med Infect Dis. 2020;36:101610. doi:10.1016/j.tmaid.2020.101610
10. Godbole GS, Day MR, Murthy S, Chattaway MA, Nair S. First report of CTX-M-15 Salmonella Typhi from England. Clin Infect Dis. 2018;66(12):1976–1977. doi:10.1093/cid/ciy032
11. Ahmad S, Tsagkaris C, Aborode AT, et al. A skeleton in the closet: the implications of COVID-19 on XDR strain of typhoid in Pakistan. Public Health Pract. 2021;2:100084. doi:10.1016/j.puhip.2021.100084
12. National Institute of Health Islamabad. Weekly field epidemiology report; 2020. Available from: https://www.nih.org.pk/wp-content/uploads/2020/09/37-FELTP-Pakistan-Weekly-Epidemiological-Report-Sep-05-12-2020.pdf.
13. World Health Organization. Drug resistant Salmonella infections in Pakistan: update. Wkly Epidemiol Monit. 2019;12:1.
14. Al Naiemi N, Zwart B, Rijnsburger MC, et al. Extended-spectrum-beta-lactamase production in a Salmonella enterica serotype Typhi strain from the Philippines. J Clin Microbiol. 2008;46(8):2794–2795. doi:10.1128/JCM.00676-08
15. Su L, Chiu C. Current system of Salmonella nomenclature used by WHO, CDC and ASM; 2005.
16. Winokur P, Canton R, Casellas J-M, Legakis N. Variations in the prevalence of strains expressing an extended-spectrum β-lactamase phenotype and characterization of isolates from Europe, the Americas, and the Western Pacific region. Clin Infect Dis. 2001;32(Supplement_2):S94–S103. doi:10.1086/320182
17. Gniadkowski M. Evolution and epidemiology of extended-spectrum β-lactamases (ESBLs) and ESBL-producing microorganisms. Clin Microbiol Infect. 2001;7(11):597–608. doi:10.1046/j.1198-743x.2001.00330.x
18. Weill F-X, Lailler R, Praud K, et al. Emergence of extended-spectrum-β-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J Clin Microbiol. 2004;42(12):5767–5773. doi:10.1128/JCM.42.12.5767-5773.2004
19. Qamar FN, Azmatullah A, Kazi AM, Khan E, Zaidi AKM. A three-year review of antimicrobial resistance of Salmonella enterica serovars Typhi and Paratyphi A in Pakistan. J Infect Dev Ctries. 2014;8(8):981–986. doi:10.3855/jidc.3817
20. Khadka S, Shrestha B, Pokhrel A, Khadka S, Joshi RD, Banjara MR. Antimicrobial resistance in Salmonella Typhi isolated from a referral Hospital of Kathmandu, Nepal. Microbiol Insights. 2021;14:11786361211056350. doi:10.1177/11786361211056350
21. Narasanna R, Chavadi M, Chandrakanth K. Prevalence of multidrug-resistant Salmonella typhi in typhoid patients and detection of blaCTX-M2 and blaCTX-M9 genes in cefetoxime-mediated extended spectrum β-lactamase-producing Salmonella typhi isolates. Biomed Res. 2018;29(14):1.
22. Zakir M, Khan M, Umar MI, Murtaza G, Ashraf M, Shamim S. Emerging trends of Multidrug-Resistant (MDR) and Extensively Drug-Resistant (XDR) Salmonella Typhi in a tertiary care hospital of Lahore, Pakistan. Microorganisms. 2021;9(12):2484. doi:10.3390/microorganisms9122484
23. Kobayashi T, Hayakawa K, Mawatari M, et al. Case report: failure under azithromycin treatment in a case of bacteremia due to Salmonella enterica Paratyphi A. BMC Infect Dis. 2014;14(1):1–4. doi:10.1186/1471-2334-14-404
24. Aziz S, Malik L. Emergence of multi-resistant enteric infection in a Paediatric unit of Karachi, Pakistan. J Pak Med Assoc. 2018;5:2–84.
25. Hooda Y, Sajib MS, Rahman H, et al. Molecular mechanism of azithromycin resistance among typhoidal Salmonella strains in Bangladesh identified through passive pediatric surveillance. PLoS Negl Trop Dis. 2019;13(11):e0007868. doi:10.1371/journal.pntd.0007868
26. Hancuh M, Walldorf J, Minta AA. Typhoid fever surveillance, incidence estimates, and progress toward typhoid conjugate vaccine introduction—worldwide, 2018–2022. MMWR Morb Mortal Wkly Rep. 2023;72:171–176. doi:10.15585/mmwr.mm7207a2
27. Clark ST, Cronin K, Corbeil AJ, Patel SN. A ten-year retrospective survey of antimicrobial susceptibility patterns among Salmonella enterica subsp. enterica serovar typhi isolates in Ontario, Canada. Microbiol Spectr. 2023;11:e04828–04822. doi:10.1128/spectrum.04828-22
28. Chen J, Ed-Dra A, Zhou H, Wu B, Zhang Y, Yue M. Antimicrobial resistance and genomic investigation of non-typhoidal Salmonella isolated from outpatients in Shaoxing city, China. Front Public Health. 2022;10:1.
29. Ain Q, Tahir M, Sadaqat A, et al. First detection of extensively drug-resistant Salmonella typhi isolates harboring VIM and GES genes for carbapenem resistance from Faisalabad, Pakistan. Microb Drug Resist. 2022;28(12):1087–1098. doi:10.1089/mdr.2022.0094
30. Rasheed F, Saeed M, Alikhan N-F, et al. Emergence of resistance to fluoroquinolones and third-generation cephalosporins in Salmonella Typhi in Lahore, Pakistan. Microorganisms. 2020;8(9):1336. doi:10.3390/microorganisms8091336
31. Haqqi A, Khurram M, Din MSU, et al. COVID‐19 and Salmonella Typhi co‐epidemics in Pakistan: a real problem. J Med Virol. 2021;93(1):184. doi:10.1002/jmv.26293
32. Khokhar F, Pickard D, Dyson Z, et al. Multiplex PCR assay to detect high risk lineages of Salmonella Typhi and Paratyphi A. PLoS One. 2022;17(7):e0267805. doi:10.1371/journal.pone.0267805
33. Ramadan AA, Abdelaziz NA, Amin MA, Aziz RK. Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended spectrum beta lactamase-producing Escherichia coli. Sci Rep. 2019;9(1):1–12. doi:10.1038/s41598-019-39730-0
34. Sajib MS, Tanmoy AM, Hooda Y, et al. Tracking the emergence of azithromycin resistance in multiple genotypes of typhoidal salmonella. Mbio. 2021;12(1):e03481–03420. doi:10.1128/mBio.03481-20
35. Nguyen MCP, Woerther P-L, Bouvet M, Andremont A, Leclercq R, Canu A. Escherichia coli as reservoir for macrolide resistance genes. Emerg Infect Dis. 2009;15(10):1648. doi:10.3201/eid1510.090696
36. Hussain A, Satti L, Hanif F, et al. Typhoidal Salmonella strains in Pakistan: an impending threat of extensively drug-resistant Salmonella Typhi. Eur J Clin Microbiol Infect Dis. 2019;38(11):2145–2149. doi:10.1007/s10096-019-03658-0
37. Fida S, Mansoor H, Saif S, Iqbal J, Khan AQ. Clinical perspectives of multiple and extensively drug-resistant typhoid; result from a tertiary care hospital from Pakistan. J Infect Dev Ctries. 2021;15(04):530–537. doi:10.3855/jidc.13539
38. Qureshi S, Naveed AB, Yousafzai MT, et al. Response of extensively drug resistant Salmonella Typhi to treatment with meropenem and azithromycin, in Pakistan. PLoS Negl Trop Dis. 2020;14(10):e0008682. doi:10.1371/journal.pntd.0008682
39. Jamilah J, Hatta M, Natzir R, et al. Analysis of existence of multidrug-resistant H58 gene in Salmonella enterica serovar Typhi isolated from typhoid fever patients in Makassar, Indonesia. New Microbes New Infect. 2020;38:100793. doi:10.1016/j.nmni.2020.100793
40. Ingle DJ, Nair S, Hartman H, et al. Informal genomic surveillance of regional distribution of Salmonella Typhi genotypes and antimicrobial resistance via returning travellers. PLoS Negl Trop Dis. 2019;13(9):e0007620. doi:10.1371/journal.pntd.0007620
41. Wang Y, Lu D, Jin Y, et al. Extensively Drug-Resistant (XDR) Salmonella Typhi outbreak by waterborne infection—Beijing municipality, China. China CDC Weekly. 2022;4(12):254. doi:10.46234/ccdcw2022.062
42. Lima NCB, Tanmoy AM, Westeel E, et al. Analysis of isolates from Bangladesh highlights multiple ways to carry resistance genes in Salmonella Typhi. BMC Genom. 2019;20(1):1–15. doi:10.1186/s12864-019-5916-6
43. Katiyar A, Sharma P, Dahiya S, Singh H, Kapil A, Kaur P. Genomic profiling of antimicrobial resistance genes in clinical isolates of Salmonella Typhi from patients infected with Typhoid fever in India. Sci Rep. 2020;10(1):1–15. doi:10.1038/s41598-020-64934-0
44. Zahid I, Sarwar A, Hussain A, Sohail M, Amin A. Antibiotyping and genotyping of extensively drug‐resistant (XDR) Salmonella sp. isolated from clinical samples of Lahore, Pakistan. J Appl Microbiol. 2022;132(1):633–641. doi:10.1111/jam.15131
45. Rahman SIA, Dyson ZA, Klemm EJ, et al. Population structure and antimicrobial resistance patterns of Salmonella Typhi isolates in urban Dhaka, Bangladesh from 2004 to 2016. PLoS Negl Trop Dis. 2020;14(2):e0008036. doi:10.1371/journal.pntd.0008036
46. da Silva KE, Tanmoy AM, Pragasam AK, et al. The international and intercontinental spread and expansion of antimicrobial-resistant Salmonella Typhi: a genomic epidemiology study. Lancet Microb. 2022;3(8):e567–e577. doi:10.1016/S2666-5247(22)00093-3
47. Gokul BN, Menezes GA, Harish BN. Acc-1 β-lactamase–producing Salmonella enterica serovar Typhi, India. Emerg Infect Dis. 2010;16(7):1170. doi:10.3201/eid1607.091643
48. Ahmed D, Hoque A, Mazumder R, et al. Salmonella enterica serovar Typhi strain producing extended-spectrum β-lactamases in Dhaka, Bangladesh. J Med Microbiol. 2012;61(7):1032–1033. doi:10.1099/jmm.0.044065-0
49. Rotimi VO, Jamal W, Pal T, Sovenned A, Albert MJ. Emergence of CTX-M-15 type extended-spectrum β-lactamase-producing Salmonella spp. in Kuwait and the United Arab Emirates. J Med Microbiol. 2008;57(7):881–886. doi:10.1099/jmm.0.47509-0
50. Jacob JJ, Pragasam AK, Vasudevan K, et al. Salmonella Typhi acquires diverse plasmids from other Enterobacteriaceae to develop cephalosporin resistance. Genomics. 2021;113(4):2171–2176. doi:10.1016/j.ygeno.2021.05.003
51. Muacevic A, Adler J, Shah A, et al. Antimicrobial Sensitivity Pattern of Salmonella Typhi: emergence of Resistant Strains. Cureus. 2021;12(11):1.
52. Marchello CS, Birkhold M, Crump JA. Complications and mortality of typhoid fever: a global systematic review and meta-analysis. J Infect. 2020;81(6):902–910. doi:10.1016/j.jinf.2020.10.030
53. Iqbal J, Dehraj IF, Carey ME, et al. A race against time: reduced azithromycin susceptibility in Salmonella enterica serovar Typhi in Pakistan. MSphere. 2020;5(4):e00215–00220. doi:10.1128/mSphere.00215-20
54. Octavia S, Chew KL, Lin RT, Teo JW. Azithromycin-resistant Salmonella enterica Serovar Typhi AcrB-R717Q/L, Singapore. Emerg Infect Dis. 2021;27(2):624. doi:10.3201/eid2702.203874
55. Duy PT, Dongol S, Giri A, et al. The emergence of azithromycin-resistant Salmonella Typhi in Nepal. JAC Antimicrob Resist. 2020;2(4):dlaa109. doi:10.1093/jacamr/dlaa109
56. Cheng M-W, Lee C-M, Wang N-Y, et al. Clinical characteristics in adult patients with Salmonella bacteremia and analysis of ciprofloxacin-nonsusceptible isolates. J Microbiol Immunol Infect. 2015;48(6):692–698. doi:10.1016/j.jmii.2015.09.001
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.