Back to Journals » Infection and Drug Resistance » Volume 13
Molecular Characterization, Drug Resistance and Virulence Analysis of Constitutive and Inducible Clindamycin Resistance Staphylococcus aureus Strains Recovered from Clinical Samples, Tehran – Iran
Authors Goudarzi M , Tayebi Z, Fazeli M, Miri M, Nasiri MJ
Received 27 February 2020
Accepted for publication 8 April 2020
Published 22 April 2020 Volume 2020:13 Pages 1155—1162
DOI https://doi.org/10.2147/IDR.S251450
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Mehdi Goudarzi,1 Zahra Tayebi,2 Maryam Fazeli,3 Mirmohammad Miri,4 Mohammad Javad Nasiri1
1Department of Microbiology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran; 2Microbiology Department, Tehran Medical Sciences Branch, Islamic Azad University, Tehran, Iran; 3Department of Virology, Pasteur Institute of Iran, Tehran, Iran; 4Department of Critical Care and Anesthesiology, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Correspondence: Mehdi Goudarzi
Department of Microbiology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran, Koodak-Yar St., Daneshjoo Blvd, Velenjak, Chamran HWY, Tehran, Iran
Tel/Fax +98 21 23872556
Email [email protected]
Background: Macrolide-lincosamide streptogramin B family is one of the important alternative antibiotics for treating staphylococcal infections. The aim of this study was to determine the characteristics and prevalence of antibiotic resistance genes in different coagulase types of clinical Staphylococcus aureus strains.
Methods: In the present study, 86 isolates with different phenotypes of MLSB resistance were investigated. In vitro susceptibility was assessed by the disk diffusion and broth microdilution methods. PCR assays were used to detect resistance-related genes. Coagulase and SCCmec types were identified by multiplex PCR assay.
Results: The prevalences of constitutive MLSB, inducible MLSB, and MS phenotypes were found to be 23%, 14.2%, and 4.9%, respectively. The rates of resistance to mupirocin, fusidic acid, and tigecycline were found to be 9.3%, 4.6%, and 2.3%, respectively. The top three predominant resistance genes were mecA, tet(M), erm(C) representing 75.6, 50, and 40.7% of isolates. mupA (7%), fusB (3.5%), and fusC (1.2%) genes were also detected among tested isolates. Coagulase types were mainly type II (34.9%), followed by III (32.6%), V (20.9%), and I (11.6%).
Conclusion: These findings indicated high resistance rate and low genetic variability with the prominence of coa type II, highlighting the particular importance of diagnosis of these strains to avoid treatment failure.
Keywords: Staphylococcus aureus, methicillin-resistant Staphylococcus aureus, polymerase chain reaction, PCR, coagulase, clindamycin
Introduction
Staphylococcus aureus is an opportunistic pathogen causing various hospital and community-acquired infections ranging from pyogenic skin infections to life-threatening diseases.1 According to the evidence, specific virulence factors such as i) cell surface components including collagen-binding protein, clumping factor, fibronectin-binding protein, and elastin-binding protein, ii) secreted factors, e.g. staphylokinase, toxic shock syndrome toxin (TST), hemolysin, exfoliative toxins, staphylococcal enterotoxins (SEs), lipase and panton-valentine leukocidin (PVL), play a pivotal role in pathogenesis and also are related to severity of the infection.1,2 Recent studies have demonstrated an evolved resistance to various antibiotic types among S. aureus, which has raised real concerns.3–6 Due to emerging simultaneous resistance to several antibacterial agents, the choice of chemotherapeutic options and treatment of serious infections caused by S. aureus has become problematic.7–9 Although various antibiotics such as vancomycin, linezolid, and quinupristin-dalfopristin may be considered as drugs of choice, S. aureus strains with reduced susceptibility and resistance to these agents also emerged. Therefore, the use of macrolide-lincosamide-streptogramin B (MLSB) antibiotics as an alternative approach to treating such infections was taken into consideration.3,4,6 However, some previous studies reported a constitutive and inducible resistance to clindamycin in S. aureus strains due to the indiscriminate use of MLSB antibiotics.4,6,10 Therefore, particular attention should be paid to the detection of constitutive and inducible clindamycin resistance genotypes and phenotypes to prevent clinical therapeutic failure in patients with S. aureus infections.11 With respect to the limited data in this context, the present study was designed to describe the phenotypic and genotypic resistance pattern and the presence of the virulence factors. Coagulase (coa) typing and staphylococcal cassette chromosome mec (SCCmec) typing were used to characterize the genotype of the constitutive and inducible clindamycin resistance isolates.
Materials and Methods
Study Design, Isolation of Bacteria and Ethics Statement
This descriptive cross-sectional study was conducted on 204 nosocomial S. aureus strains recovered from clinical samples such as pus (36.3%), wound (31.9%), blood (13.7%), sputum (7.3%), cerebrospinal fluid (5.9%), and urine (4.9%) during the period of 1 year, from August 2018 to July 2019. The Ethics Committee of the Shahid Beheshti University of Medical Sciences in Tehran, Iran certified the protocol of this project (IR. SBMU. MSP.REC. 1396.700). At first, S. aureus isolates were phenotypically identified using standard microbiological and biochemical techniques and then were subjected to polymerase chain reaction (PCR) assay for the presence of nuc gene for definitive confirmation.12 A total of 86 nosocomial S. aureus isolates were included in the study based on resistance to erythromycin and resistance and/or susceptibility to clindamycin in accordance with standard clinical and laboratory standard institute (CLSI) guidelines. Isolates with resistance to both clindamycin and erythromycin-resistant were considered to be constitutive resistance phenotype (cMLSB). Isolates with resistance to erythromycin but susceptible to clindamycin were tested by the D test. Inducible resistance phenotype (iMLSB) was defined for isolates showing resistance to erythromycin and susceptible to clindamycin with a D-shaped zone around the clindamycin disk, flattened from the side of erythromycin disk. Isolates with both inhibition zones showing a circular shape (D test negative) were classified as the MS phenotype (CLSI 2019).
Antimicrobial Activities
The disk diffusion method using cefoxitin (30 μg) disk in Mueller-Hinton agar (Merck, Germany) according to the CLSI was applied for the screening of methicillin resistance isolates. In addition, PCR assay was used for the detection of mecA gene.12
The Kirby–Bauer disk diffusion method was used to determine the susceptibility of the isolates against penicillin, ceftriaxone, amikacin, gentamicin, tobramycin, kanamycin, tetracycline, linezolid, teicoplanin, ciprofloxacin, rifampicin, quinupristin-dalfopristin, and trimethoprim-sulfamethoxazole (Mast Co., UK) based on the CLSI recommendation (CLSI 2019). The minimum inhibitory concentration (MIC) value for vancomycin, mupirocin, tigecycline, and fusidic acid was determined using the broth microdilution method. Results for fusidic acid and tigecycline were interpreted according to the European Committee for antimicrobial susceptibility testing (EUCAST) breakpoints (http://www.eucast.org). Low-level and high-level mupirocin resistance (LLMUPR, HLMUPR) were defined if MIC values of 8–256 µg/mL and ≥512 µg/mL were obtained. S. aureus strains ATCC 25923, ATCC 43300 and ATCC 29213 were used as reference strains.
DNA Extraction
In this study, the DNA of each strain was extracted using the phenol-chloroform extraction method with the modification of adding lysostaphin (Sigma-Aldrich, United States) for bacterial lysis. DNA concentration and purity were investigated using a NanoDrop 2000 spectrophotometer (spectrophotometer, Thermo Scientific, Wilmington, DE, USA).
Amplification of Resistance-Related Genes
The resistance encoding genes vanA, vanB, mupB, mupA, fusA, fusB, fusC, mecC, msr(A), msr(B), erm(A), erm(B), erm(C), tet(M), ant (4΄)-Ia, aac (6΄)-Ie/aph (2˝), aph (3΄)-IIIa and virulence determinants including exfoliative toxin (eta, and etb), Panton-Valentine leukotoxin (pvl), and toxic shock syndrome toxin (tst) genes were detected by PCR.12,13 In order to detect target genes, PCR amplified products were resolved using electrophoresis in 1.8% (w/v) agarose gel, stained with ethidium bromide (0.5 mg/mL) and visualized under UV light using a gel documentation system (Bio-Rad Laboratories, USA).
SCCmec Typing
For methicillin-resistant S. aureus (MRSA) strains, determination of SCCmec types was performed using multiplex PCR assay, as previously described by Nezhad et al S. aureus ATCC 10442 (type I), S. aureus N315 (type II), S. aureus 85/2082 (type III), S. aureus MW2 (type IV), S. aureus WIS 173 (type V) and S. aureus HDE288 (type VI) were recruited as control strains.
Staphylocoagulase (SC) Typing
Multiplex PCR assay with four-set primers (A-D), was used for determination of SC types (I–X) as previously described by Hirose et al Set A contained primers for identification of SC types I, II, III, IVa, IVb, Va, and VI while set B contained primers for identifying SC types VII, VIII, and X. Set C was used to identify SC types IX and Vb. SC types IVa and IVb were distinguished using set four primers (Set D).14
Results
Out of 204 S. aureus tested isolates, the overall prevalence of cMLSB, iMLSB, and MS phenotypes were 47 (23%), 29 (14.2%), and 10 (4.9%), respectively. Out of 86 tested isolates, 25 isolates were obtained from hospital H1 (29.1%), 20 isolates from hospital H2 (23.3%), 18 isolates from hospital H3 (20.9%), and 23 isolates from hospital H4 (26.7%). The under-study isolates were recovered from wound (28/86, 32.6%), pus (21/86, 24.4%), blood (19/86, 22.1%), sputum (7/86, 8.1%), CSF (6/86, 7%), and urine (5/86, 5.8%). According to our analysis, the rate of invasive and non-invasive S. aureus was found to be 29.1% and 70.9%, respectively. All the invasive S. aureus isolates were methicillin-resistant with iMLSB (40%; 10/25), MS (40%; 10/25) and cMLSB (20%; 5/25) phenotypes. A total of 86 S. aureus strains included in present study, 47 were isolated from female patients (54.7%) and 39 were recovered male patients (45.3%) with a median age of 41.4 years, ranging from 15 to 59 years. Of these examined 86 S. aureus strains, 75.6% (65/86) and 24.4% (21/86) were MRSA and methicillin-sensitive S. aureus (MSSA), respectively. The cMLSB phenotype was observed in both MRSA (26, 30.2%) and MSSA (21, 24.4%) strains, whereas phenotypes of iMLSB and MS were found only in MRSA strains. The most common S. aureus isolates with iMLSB phenotype in the present study were isolated from wound infection (19.8%, 17/86) while cMLSB phenotypes were from pus (22.1%, 19/86) and MS phenotypes were from blood infection (9.3%, 8/86). Distribution of different MLSB phenotypes in S. aureus strains isolated from various clinical specimens is shown in Table 1. Based on data obtained from the disk diffusion test, all isolated S. aureus strains were found to be susceptible to linezolid, teicoplanin, and vancomycin. As summarized in Table 2, resistance to all antibacterial agents (but no amikacin and fusidic acid) was more common among MRSA isolates than among MSSA isolates. All the four of the fusidic acid-resistant isolates were MSSA isolates with cMLSB phenotype, based on the results of the micro-broth dilution method. The rate of resistance to mupirocin was found to be 9.3%. Six isolates indicating high-level resistance to mupirocin (HLMUPR) belonged to MRSA strains with iMLSB (5 isolates) and MS (one isolate) phenotypes and two isolates showing low-level resistance to mupirocin (LLMUPR) were identified as MRSA strains with iMLSB phenotype. Two tigecycline resistant isolates belonged to two MRSA strains with iMLSB and MS phenotypes.
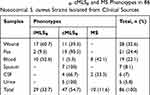 |
Table 1 Distribution of iMLSB, cMLSB and MS Phenotypes in 86 Nosocomial S. aureus Strains Isolated from Clinical Sources |
 |
Table 2 Antimicrobial Resistance Pattern of Nosocomial MRSA and MSSA Isolates with Inducible and Constitutive Phenotype |
Our findings indicated that all isolates were typed using the SC typing method. These isolates were distinguished into 4 types of SC. The predominant SC type was II (34.9%, 30/86), followed by III (32.6%, 28/86), V (20.9%, 18/86), and I (11.6%, 10/86). All the MSSA isolates belonged to SC type III. All MS phenotype MRSA isolates belonged to SC type I. Among iMLSB phenotype MSRA isolates, the most predominant SC types were II and V representing 25.6% (22/86) and 8.1% (7/86) of isolates. cMLSB phenotypes were distributed in SC types III, II, and V accounting for 32.6%, 9.3%, and 12.8%, respectively.
Resistance encoding genes analysis showed that the most prevalent gene was mecA (75.6%, 65/86), followed by tet(M) (50%, 43/86), erm(C) (40.7%, 35/86), ant (4΄)-Ia (29.1%, 25/86), aac (6΄)-Ie/aph (2˝) (20.9%, 18/86), msr(A) (20.9%, 18/86), aph (3΄)-IIIa (14%, 12/86), erm(B) (14%, 12/86), msr(B) (9.3%, 8/86), erm(A) (8.1%, 7/86), mupA (7%, 6/86), fusB (3.5%, 3/86), and fusC (1.2%, 1/86). Our findings showed that no PCR products were found for the resistance genes vanA, vanB, mupB, fusA, and mecC. The distribution of SCCmec types in the 65 MRSA isolates showed that SCCmec type III was the most prevalent type found in 33 isolates (50.8%), followed by type II in 20 isolates (30.8%), and type IV in 12 isolates (18.4%). SCCmec types I and V were not found in our isolates. The resistance profile and distribution of coa and SCCmec types in MRSA and MSSA isolates with inducible and constitutive phenotype are presented in Table 3.
 |
Table 3 Distribution of coa, SCCmec Types and Resistance Profiles of Nosocomial MRSA and MSSA Isolates with Inducible and Constitutive Phenotype |
Discussion
Little is known about the emergence, distribution and molecular types of constitutive and inducible clindamycin resistance S. aureus strains in Iran. Accordingly, our study focused on the identification of molecular characteristics and understanding of cMLSB, iMLSB S. aureus isolates epidemiology. Our research highlighted several new findings in relation to MRSA and MSSA isolates with inducible and constitutive resistance phenotype including a relatively high prevalence of inducible and constitutive resistance and distinct molecular types with genetic diversity. According to the evidence, the prevalence rate of iMLSB phenotype among S. aureus isolates was markedly varied across the geographical region and among health-care settings. The current finding showed a prevalence rate of 14.2% for iMLSB which is higher than the reported rate in Nepal (11.48%),3 Iran (8.6%),5 Egypt (7.7%),15 Turkey (7.8%)4 and lower than those reported in India (37.5%).16 However, the results of previous studies conducted in Iran noted significant variation in iMLSB prevalence rates in different areas10,17 ranging from 6% to 32.3%. This work presented a relatively high prevalence of iMLSB phenotype, which highlighted the need to prescribe of macrolides in a logical manner, in order to change in resistance pattern. However, the true evaluation of iMLSB S. aureus isolates prevalence depends on the accurate diagnosis, geographical variation, characteristic of health-care setting, and population under study.
In the current research, the prevalence of cMLSB among S. aureus was found to be 23%, which was similar to reported rate, by Delialioglu et al (24.3%),4 and Eksi (20.4%).18 However, a lower and higher percentage of cMLSB were also reported in previous studies performed by Khashei et al (82.9%),5 Adhikari et al (29.25%),3 Mansouri et al (28.4%),19 Sedaghat et al (32.1%),17 and Sasirekha et al (13.1%).6 These variations in the prevalence of cMLSB among S. aureus in different parts of the world could be attributed to the difference in consumption of macrolides in community and hospital settings, study design, population and geographical distribution, and the spread of specific molecular types. As presented in Table 2, the cMLSB phenotype was higher in MRSA (30.2%) as compared to MSSA (24.4%) and iMLSB and MS phenotypes were found only in MRSA strains, which is consistent with previous reports from Nepal,3 Iran,19 Egypt,15 and Turkey.4
In our study, low resistance rate to mupirocin (9.3%) was noted which is in agreement with other studies conducted in Iran (6%),7 India (5%),8 and Jordan (2.6%).20 Furthermore, the findings of the present study demonstrate that 7% of tested isolates were found to have resistance to mupirocin at a high level which are quite similar to the results of a study conducted by Liu and colleagues in China. They reported a prevalence rate of 6.6% for isolates with high-level mupirocin-resistant.21 However, there has been a higher prevalence of HLMUPR strains in Iran (25%)22 and Egypt (61.5%).23 Different results of these studies may be due to study design (patient characteristics and specimen types), specific type dissemination among patients and unrestricted policies in taking mupirocin. This study showed that all HLMUPR isolates were mupA-positive (7%). González-Domínguez et al (27.2%)24 and Abbasi-Montazeri et al (34%)7 reported a higher percentage of mupA. Shahsavan et al reported that mupA was responsible for the resistance to mupirocin only in MRSA strains with cMLSB phenotype. Conversely, in the present research, this gene was present in MRSA strains with iMLSB and MS resistance phenotype.
Tigecycline is a reliable treatment option against many infections caused by MDR isolates especially MRSA. So far, there have been few reports published on the emergence of S. aureus strains with reduced susceptibility and resistance to tigecycline. We found the low numbers of tigecycline resistant isolates (2.3%). However, different resistance rates to tigecycline among S. aureus isolates are reported in Libya (3.6%),25 Turkey (2%),26 and Iran (6.6%).9
Recently published data from Asian countries indicated a low prevalence of resistance to fusidic acid (<10%).27,28 We found a low prevalence (4.7%) of resistance to fusidic acid among our isolates carrying fusB (3 isolates), and fusC (1 isolate) genes. This observation is consistent with data from a recent multicenter study in Iran that showed a low prevalence of MRSA fusidic acid-resistant (3%) among 726 studied S. aureus isolates.28 Various resistance rates to fusidic acid have been described in many countries such as Greece (62.4%), Ireland (19.9%), Australia (7.0%), Canada (7.0%), and the United States (0.3%).29 Notably, this study showed that fusidic acid resistance was only seen among MSSA isolates which was in contrast to a report from China that indicated the prevalence of fusidic acid resistance among MRSA isolates was higher significantly than that among MSSA isolates.30 Yu and colleagues also reported a 10.5% incidence of fusB genes while fusC and fusA genes were not detected in any of the isolates examined.30 This indicates that fusB is the predominant determinant responsible for resistance to fusidic acid among MSSA isolates with cMSLB resistance phenotype in Iran.
ant (4΄)-Ia as the most prevalent aminoglycoside resistance gene was present in 25 strains (29.1%) which was higher than those reported in Turkey (24%)31 and lower than that reported rate in India (9%).32 However, much higher rates have also been reported by Nezhad et al (94.7%)12 and Ida et al (84.5%).33 In our research, the prevalence of aac (6΄)-Ie/aph (2˝) as the second commonly detected aminoglycoside resistance gene was found to be 20.9% which is lower than that reported by Ardic et al (60.5%).31 Akpaka et al reported that in MRSA strains, 20% harbored ant (4΄)-Ia gene.34 Many reports from different parts of Asia such as Turkey (8%),31 and India (9%)34 have shown a low prevalence of the ant (4ʹ)-Ia gene. Likewise, in the current work, 14% of isolates were found to carry aph (3΄)-IIIa. This variation in aminoglycoside resistance determinant frequency displayed that factors such as study design, specimen types, different policies in aminoglycosides consumption, and horizontal gene transfer among the strains may be involved.
Based on the literature, inducible and constitutive resistance in S. aureus strains is mediated by both erm and msr genes. Our analysis recorded erm(C) (40.7%) and erm(A) (8.1%) as the highest and lowest erythromycin resistance gene. Although there is a discrepancy in the prevalence rate of erm(C) gene in different parts of the world, a similar finding from Iran has also been reported.17 These findings are in concordance with those described by Fasihi et al, which reported prevalence of erm(C) and erm(A) genes to be 20.5% and 11% for MRSA strains.35 In accordance with our results, Schmitz and colleagues reported that the erm(A) gene was more common in MRSA isolates compared to MSSA isolates (88% vs 38%) and occurred mainly in cMLSB expression strains.36 A high percentage of msr(A) (20.9%) was obtained in the current research, similar to a study conducted by Sedaghat et al (43.6%),17 Nezhad et al (47.3%).12 In accordance with several investigators who reported the prevalence of erm(B) gene at a low level, we detected erm(B) gene in 5.8% and 8.1% inducible and constitutive resistant strains, respectively. A study from Texas also showed a high frequency of erm(B) gene in inducible resistant strains (46.3%).37 This discrepancy could be because of the distribution of some specific clonal lineages in communities and hospitals and might be more closely related to the usage of particular macrolides and ketolides in our health-care settings.
Our data related to SCCmec types are in concordance with many studies that demonstrated SCCmec types I, II, III are related to hospital-acquired S. aureus infections while IV and V are prominent types in community-acquired S. aureus infections.12,21,24 We confirmed SCCmec type IV in MRSA isolates with cMLSB (5.8%) and iMLSB (8.1%) phenotype. This finding supports a shift in these isolates from our community to hospital.
According to the coa typing results, predominant SC type was II (34.9%), followed by III (32.6%), V (20.9%), and I (11.6%). This was in comparison to the previous report by Hirose et al in Japan which indicated coa type II, VII and I accounted for 91.9%, 3.9% and 1.7% of isolates.14 We detected 2 SC types (II–V) among iMLSB S. aureus strains suggesting the clonal distribution of tested isolates in this region of Iran. In research involving 157 S. aureus strains from clinical specimens, nine different patterns of coa gene have been detected.38 In another study from Thailand on 129 MRSA isolates from 17 hospitals, Janwithayanuchit et al determined four different genotypes from coagulase gene typing of tested strains.39 They showed that the most prevalent coa type was III (82.2%) followed by IV (14%), II (2.3%), and I (1.5%). The analysis was carried out by Younis Omar et al on 75 MRSA isolated from different ICUs grouped into three different types based on the polymorphism of coa gene products by PCR.40 The similarity between coa genes of examined strains highlighted that it may be as a predictor for specific inducible resistant S. aureus strains.
Conclusion
Given the presence of various types of MLSB resistance in our survey, special attention should be given to diagnosing these resistance types in order to judicious use of clindamycin. However, some resistance patterns were related to certain SCCmec and coa types. These strains have low genetic variability with a predominance of coa type II and SCCmec type III. Further researches should be performed in other regions of Iran to keep track of the emerging coa types.
Funding
This work was supported by the fund in the Research Deputy of Shahid Beheshti University of Medical Sciences, Tehran, Iran (Grant No. 12688).
Disclosure
The authors declare that they have no conflict interest.
References
1. Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–661. doi:10.1128/CMR.00134-14
2. Vandenesch F, Lina G, Henry T. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: a redundant arsenal of membrane-damaging virulence factors? Front Cell Infect Microbiol. 2012;2:12. doi:10.3389/fcimb.2012.00012
3. Adhikari R, Shrestha S, Barakoti A, Amatya R. Inducible clindamycin and methicillin resistant Staphylococcus aureus in a tertiary care hospital, Kathmandu, Nepal. BMC Infect Dis. 2017;17(1):483. doi:10.1186/s12879-017-2584-5
4. Delialioglu N, Aslan G, Ozturk C, Baki V, Sen S, Emekdas G. Inducible clindamycin resistance in staphylococci isolated from clinical samples. Jpn J Infect Dis. 2005;58:104–106.
5. Khashei R, Malekzadegan Y, Ebrahim-Saraie HS, Razavi Z. Phenotypic and genotypic characterization of macrolide, lincosamide and streptogramin B resistance among clinical isolates of staphylococci in southwest of Iran. BMC Res Notes. 2018;11(1):1–6. doi:10.1186/s13104-018-3817-4
6. Sasirekha B, Usha M, Amruta J, Ankit S, Brinda N, Divya R. Incidence of constitutive and inducible clindamycin resistance among hospital-associated Staphylococcus aureus. 3 Biotech. 2014;4(1):85–89. doi:10.1007/s13205-013-0133-5
7. Abbasi-Montazeri E, Khosravi AD, Feizabadi MM, et al. The prevalence of methicillin resistant Staphylococcus aureus (MRSA) isolates with high-level mupirocin resistance from patients and personnel in a burn center. Burns. 2013;39(4):650–654. doi:10.1016/j.burns.2013.02.005
8. Gadepalli R, Dhawan B, Mohanty S, et al. Mupirocin resistance in Staphylococcus aureus in an Indian hospital. Diagn Microbiol Infect Dis. 2007;58(1):125–127. doi:10.1016/j.diagmicrobio.2006.10.012
9. Yousefi M, Fallah F, Arshadi M, Pourmand M, Hashemi A, Pourmand G. Identification of tigecycline-and vancomycin-resistant Staphylococcus aureus strains among patients with urinary tract infection in Iran. New Microbes New Infect. 2017;19:8–12. doi:10.1016/j.nmni.2017.05.009
10. Moosavian M, Shoja S, Rostami S, Torabipour M, Farshadzadeh Z. Inducible clindamycin resistance in clinical isolates of Staphylococcus aureus due to erm genes, Iran. Iran J Microbiol. 2014;6(5):421.
11. Lewis JS, Jorgensen JH. Inducible clindamycin resistance in staphylococci: should clinicians and microbiologists be concerned? Clin Infect Dis. 2005;40(2):280–285. doi:10.1086/426894
12. Nezhad RR, Meybodi SM, Rezaee R, Goudarzi M, Fazeli M. Molecular characterization and resistance profile of methicillin resistant Staphylococcus aureus strains isolated from hospitalized patients in intensive care unit, Tehran-Iran. Jundishapur J Microbiol. 2017;10.
13. Castanheira M, Watters AA, Bell JM, Turnidge JD, Jones RN. Fusidic acid resistance rates and prevalence of resistance mechanisms among Staphylococcus spp. isolated in North America and Australia, 2007-2008. Antimicrob Agents Chemother. 2010;54:3614–3617. doi:10.1128/AAC.01390-09
14. Hirose M, Kobayashi N, Ghosh S, et al. Identification of Staphylocoagulase genotypes I-X and discrimination of Type IV and V subtypes by multiplex PCR assay for clinical isolates of Staphylococcus aureus. Jpn J InfectDis. 2010;63:257–263.
15. Kilany AA. Inducible clindamycin resistance among clinical isolates of Staphylococcus aureus. Menoufia Med J. 2016;29:228. doi:10.4103/1110-2098.192418
16. Lall M, Sahni A. Prevalence of inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. Med J Armed Forces India. 2014;70:43–47. doi:10.1016/j.mjafi.2013.01.004
17. Sedaghat H, Esfahani BN, Mobasherizadeh S, et al. Phenotypic and genotypic characterization of macrolide resistance among Staphylococcus aureus isolates in Isfahan, Iran. Iran J Microbiol. 2017;9:264.
18. Eksi F, Gayyurhan ED, Bayram A, Karsligil T. Determination of antimicrobial susceptibility patterns and inducible clindamycin resistance in Staphylococcus aureus strains recovered from southeastern Turkey. J Microbiol Immunol Infect. 2011;44(1):57–62. doi:10.1016/j.jmii.2011.01.011
19. Mansouri S, Sadeghi J. Inducible clindamycin resistance in methicillin-resistant and-susceptible Staphylococcus aureus isolated from south east of Iran. Jundishapur J Microbiol. 2014;7.
20. Aqel A, Ibrahim A, Shehabi A. Rare occurrence of mupirocin resistance among clinical Staphylococcus in Jordan. Acta Microbiol Imm H. 2012;59(2):239–247. doi:10.1556/AMicr.59.2012.2.8
21. Liu Q-Z, Wu Q, Zhang Y-B, et al. Prevalence of clinical meticillin-resistant Staphylococcus aureus (MRSA) with high-level mupirocin resistance in Shanghai and Wenzhou, China. Int J Antimicrob Agents. 2010;35:114–118. doi:10.1016/j.ijantimicag.2009.09.018
22. Shahsavan S, Emaneini M, Khoshgnab BN, et al. A high prevalence of mupirocin and macrolide resistance determinant among Staphylococcus aureus strains isolated from burnt patients. Burns. 2012;38:378–382. doi:10.1016/j.burns.2011.09.004
23. Barakat GI, Nabil YM. Correlation of mupirocin resistance with biofilm production in methicillin-resistant Staphylococcus aureus from surgical site infections in a tertiary centre, Egypt. J Glob Antimicrob Resist. 2016;4:16–20. doi:10.1016/j.jgar.2015.11.010
24. González-Domínguez M, Seral C, Potel C, et al. Genotypic and phenotypic characterization of methicillin-resistant Staphylococcus aureus (MRSA) clones with high-level mupirocin resistance. Diagn Microbiol Infect Dis. 2016;85(2):213–217. doi:10.1016/j.diagmicrobio.2016.02.021
25. Zorgani A, Elahmer O, Ziglam H, Ghenghesh KS. In-vitro activity of tigecycline against methicillin-resistant Staphylococcus aureus isolated from wounds of burn patients in Tripoli-Libya. J Microbiol Infect D. 2012;2(3):109–112. doi:10.5799/ahinjs.02.2012.03.0053
26. Oksuz L, Gurler N. Susceptibility of clinical methicillin-resistant Staphylococci isolates to new antibiotics. J Infect Dev Ctries. 2013;7(11):825–831. doi:10.3855/jidc.3867
27. Wang J-L, Tang H-J, Hsieh P-H, et al. Fusidic acid for the treatment of bone and joint infections caused by meticillin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2012;40:103–107. doi:10.1016/j.ijantimicag.2012.03.010
28. Rahimi F, Bouzari M, Katouli M, Pourshafie MR. Antibiotic resistance pattern of methicillin resistant and methicillin sensitive Staphylococcus aureus isolates in Tehran, Iran. Jundishapur J Microbiol. 2013;6(2):144–149. doi:10.5812/jjm.4896
29. Deotale V, Mendiratta D, Raut U, Narang P. Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. Indian J Med. Microbiol. 2010;28(2):124. doi:10.4103/0255-0857.62488
30. Yu F, Liu Y, Lu C, et al. Dissemination of fusidic acid resistance among Staphylococcus aureus clinical isolates. BMC Microbiol. 2015;15:210. doi:10.1186/s12866-015-0552-z
31. Ardic N, Sareyyupoglu B, Ozyurt M, Haznedaroglu T, Ilga U. Investigation of aminoglycoside modifying enzyme genes in methicillin-resistant staphylococci. Microbiol Res. 2006;161(1):49–54. doi:10.1016/j.micres.2005.05.002
32. Perumal N, Murugesan S, Krishnan P. Distribution of genes encoding aminoglycoside-modifying enzymes among clinical isolates of methicillin-resistant staphylococci. Indian J Med Microbiol. 2016;34(3):350. doi:10.4103/0255-0857.188339
33. Ida T, Okamoto R, Shimauchi C, Okubo T, Kuga A, Inoue M. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J Clin Microbiol. 2001;39(9):3115–3121. doi:10.1128/JCM.39.9.3115-3121.2001
34. Akpaka PE, Roberts R, Monecke S. Molecular characterization of antimicrobial resistance genes against Staphylococcus aureus isolates from Trinidad and Tobago. J Infect Public Health. 2017;10(3):316–323. doi:10.1016/j.jiph.2016.05.010
35. Fasihi Y, Saffari F, Ghahraman MRK, Kalantar-Neyestanaki D. Molecular detection of macrolide and lincosamide-resistance genes in clinical methicillin-resistant Staphylococcus aureus isolates from Kerman, Iran. Arch Pediatr Infect Dis. 2017;5.
36. Schmitz F-J, Sadurski R, Kray A, et al. Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J Antimicrob Chemother. 2000;45(6):891–894. doi:10.1093/jac/45.6.891
37. Chavez-Bueno S, Bozdogan B, Katz K, et al. Inducible clindamycin resistance and molecular epidemiologic trends of pediatric community-acquired methicillin-resistant Staphylococcus aureus in Dallas, Texas. Antimicrob Agents Chemother. 2005;49(6):2283–2288. doi:10.1128/AAC.49.6.2283-2288.2005
38. Afrough P, Pourmand MR, Sarajian AA, Saki M, Saremy S. Molecular investigation of Staphylococcus aureus, coa and spa genes in Ahvaz hospitals, staff nose compared with patients clinical samples. Jundishapur J Microbiol. 2013;6:1O.
39. Janwithayanuchit I, Ngam-Ululert S, Paungmoung P, Rangsipanuratn W. Epidemiologic study of methicillin-resistant Staphylococcus aureus by coagulase gene polymorphism. Sci Asia. 2006;32:127–132. doi:10.2306/scienceasia1513-1874.2006.32.127
40. Omar NY, Ali HAS, Harfoush RAH, El Khayat EH. Molecular typing of methicillin resistant Staphylococcus aureus clinical isolates on the basis of protein A and coagulase gene polymorphisms. Int J Microbiol. 2014;2014.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
