Back to Journals » Infection and Drug Resistance » Volume 14
Molecular and Functional Characterization of a Novel Plasmid-Borne blaNDM-Like Gene, blaAFM-1, in a Clinical Strain of Aeromonas hydrophila
Authors Lin X, Lu J, Qian C, Lin H, Li Q, Zhang X, Liu H, Sun Z, Zhou D, Lu W, Zhu M, Zhang H, Xu T, Li K, Bao Q, Lin L
Received 22 December 2020
Accepted for publication 31 March 2021
Published 22 April 2021 Volume 2021:14 Pages 1613—1622
DOI https://doi.org/10.2147/IDR.S297419
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Xi Lin,1,2 Junwan Lu,1,2 Changrui Qian,1,2 Hailong Lin,1– 3 Qiaoling Li,1– 3 Xueya Zhang,1– 3 Hongmao Liu,1– 3 Zhewei Sun,1,2 Danying Zhou,1,2 Wei Lu,1,2 Mei Zhu,4 Hailin Zhang,2,3 Teng Xu,5 Kewei Li,1,2 Qiyu Bao,1– 3 Li Lin2,3
1Key Laboratory of Medical Genetics of Zhejiang Province, Key Laboratory of Laboratory Medicine, Ministry of Education of China, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, 325035, People’s Republic of China; 2Institute of Biomedical Informatics, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, 325035, People’s Republic of China; 3The Second Affiliated Hospital and Children’s Hospital, Wenzhou Medical University, Wenzhou, 325027, People’s Republic of China; 4Department of Clinical Laboratory, Zhejiang Hospital, Hangzhou, Zhejiang, 310013, People’s Republic of China; 5Institute of Translational Medicine, Baotou Central Hospital, Baotou, 014040, People’s Republic of China
Correspondence: : Li Lin
The Second Affiliated Hospital and Yuying Children’s Hospital, Wenzhou Medical University, Wenzhou, 325027, People’s Republic of China
Tel +86-577-88002134
Email [email protected]
Qiyu Bao
Institute of Biomedical Informatics, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, 325035, People’s Republic of China
Tel/Fax +86-577-86699398
Email [email protected]
Purpose: An increasing frequency of antibiotic resistance has been observed in both clinical and environmental Aeromonas hydrophila isolates in recent years. However, there are still very few in-depth studies regarding the role of plasmids in the antibiotic resistance of A. hydrophila. Hence, we investigated the molecular and functional characterization of a multidrug-resistant plasmid encoding an NDM-like metallo-β-lactamase, AFM-1, in the clinical A. hydrophila isolate SS332.
Methods: The minimum inhibitory concentrations (MICs) of 24 antibiotics against A. hydrophila SS332 were measured by the agar dilution method. The genome of A. hydrophila SS332 was sequenced with PacBio and Illumina platforms. Six plasmid-borne antimicrobial resistance genes were chosen for cloning, including blaAFM-1, blaOXA-1, msr(E), mph(E), aac(6ʹ)-Ib10, and aph(3ʹ)-Ia. Phylogenetic analysis, amino acid sequence alignment, and comparative genomic analysis were performed to elucidate the active site requirements and genetic context of the blaAFM-1 gene.
Results: A. hydrophila SS332 showed high levels of resistance to 15 antibiotics, especially those with MIC levels at or above 1024 μg/mL, including ampicillin, cefazolin, ceftriaxone, aztreonam, spectinomycin, and roxithromycin. Six plasmid-borne resistance genes from A. hydrophila were verified to be functional in E. coli DH5α. AFM-1 shared 86% amino acid identity with NDM-1 and showed resistance to ampicillin, cefazolin, cefoxitin, and ceftazidime. In addition, the blaAFM-1 gene was associated with three different novel ISCR19-like elements, designated ISCR19-1, ISCR19-2 and ∆ISCR19-3, which may be involved in the acquisition and mobilization of the blaAFM-1 gene.
Conclusion: Our investigation showed that plasmid-borne resistance genes can contribute to antibiotic resistance in A. hydrophila SS332. A novel blaNDM-like gene, blaAFM-1, was verified to be functional and associated with novel ISCR19-like elements. This fact indicated the risk of spread of blaAFM-1 genes and ISCR19-like elements.
Keywords: Aeromonas hydrophila, whole-genome sequencing, plasmid-borne resistance genes, blaAFM-1, ISCR19-like elements
Introduction
Aeromonas hydrophila is a gram-negative bacterium that is ubiquitous in aquatic environments and can cause infections in fish,1 amphibians,2 reptiles,3 and humans.4 It has been identified as an antibiotic-resistant and virulent etiologic agent in an increasing list of human diseases, including gastroenteritis,5 diarrhea,6 necrotizing fasciitis,7 septicemia,8 meningitis,9 and hemolytic uremic syndrome.10 Unfortunately, clinical and environmental isolates of A. hydrophila are becoming increasingly resistant to multiple antibiotic agents, including resistance to colistin and carbapenem, which are considered as the last line of defense against multidrug-resistant infections.
Hughes et al reported that an A. hydrophila AHNIH1 carrying the blaKPC-2 gene isolated from perirectal surveillance culture was resistant to ertapenem.11 Similarly, A. hydrophila GSH8-2 carrying the blaKPC-2 gene with reduced susceptibility to all tested β-lactams was isolated from wastewater treatment plant effluent in Japan.12 Interestingly, the abovementioned antimicrobial resistance genes (ARGs) are all located on plasmids in A. hydrophila.12
Antimicrobial resistance is a global public health challenge that threatens human health. The increasing incidence of phenotypic resistance to antibiotics is mainly attributed to abuse of antimicrobials and acquisition of ARGs by bacterial pathogens.13 Several phenotypic properties of bacteria have been proven to be encoded on plasmids, such as antimicrobial resistance and virulence factors.14 In addition, the dissemination of ARGs is closely associated with mobile genetic elements, including plasmids, which increase their ability to replicate and contribute to self-transmission or horizontal gene transfer among different bacterial species. It has been reported that A. hydrophila is able to harbor one or more plasmids. However, there are still very few in-depth studies regarding the role of plasmids in the antibiotic resistance of A. hydrophila.
In this study, the multidrug-resistant clinical A. hydrophila strain SS332 was isolated from a fecal sample from a male gastric carcinoma patient in Lishui Central Hospital, Zhejiang, China. Genome sequencing showed that the plasmid pSS332-218k from SS332 carries 12 putative ARGs, including blaAFM-1, which is a blaNDM-like gene with 86% identity to blaNDM-14. The metallo-β-lactamase gene blaAFM-1 was first cloned, and its antibiotic resistance profile was verified. Phylogenetic analysis, amino acid sequence alignment, and comparative genomic analysis were performed to elucidate the active site requirements and genetic context of the blaAFM-1 gene.
Materials and Methods
Bacterial Strains and Plasmids
A. hydrophila SS332 was isolated from a fecal specimen from a 76-year-old male patient in Lishui Central Hospital, Zhejiang, China in 2013. It was identified by a microorganism autoanalysis system (VITEK®2, BioMérieux, France), and homologous comparisons of the 16S rRNA gene sequence to the GenBank database were performed by using the BLAST program (www.ncbi.nlm.nih.gov/BLAST/). Escherichia coli ATCC 25922 was used as the quality control strain in antimicrobial susceptibility testing. E. coli strain DH5α was used as a recipient for cloning experiments. pUCP20 and pUCP24 were used as vectors in cloning experiments.
Antimicrobial Susceptibility Testing
The agar dilution method was employed to measure the minimum inhibitory concentration (MIC) of antibiotics against A. hydrophila SS332 according to the Clinical and Laboratory Standards Institute (CLSI) guidelines, M45 3rd edition (2015). Twelve β-lactam antibiotics (ampicillin, cefminox, cefazolin, cefoxitin, ceftazidime, cefotaxime, ceftriaxone, cefoselis, cefepime, aztreonam, imipenem, and meropenem), four aminoglycoside antibiotics (spectinomycin, gentamicin, kanamycin, and amikacin), two macrolide antibiotics (erythromycin and roxithromycin), two phenicol antibiotics (chloramphenicol and florfenicol), three quinolone antibiotics (nalidixic, ciprofloxacin and levofloxacin) and polymyxin B were tested. E. coli ATCC 25922 was used as the quality control strain. The results were interpreted according to the standards of the CLSI.
Whole-Genome Sequencing and Bioinformatic Analysis
Genomic DNA of A. hydrophila SS332 was extracted by using a Bacterial Genomic DNA Miniprep kit (Generay, Shanghai, China) following the manufacturer’s instructions. Whole-genome sequencing was performed with Illumina (HiSeq 2500, Illumina, United States) and PacBio (PacBio RS II, Pacific Biosciences, United States) platforms. The initial complete genome was assembled from PacBio long reads by Canu v1.7 and then corrected by Pilon with Illumina short reads. Potential open reading frames (ORFs) were predicted using Prodigal and annotated against the UniProt/Swiss-Prot and nonredundant protein databases using the BLASTX program. Annotation of ARGs and mobile genetic elements was performed using online databases, including the Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/) and ISfinder (https://www-is.biotoul.fr/). Similar sequences of pSS332-218k showing >50% query coverage were chosen for comparative genomic analysis in the nr/nt database with BLASTN. Amino acid alignment for AFM-1 and representative subclass B1 metallo-β-lactamase (MBLs) was performed using the program Clustal W (https://www.genome.jp/tools-bin/clustalw), and the final output was produced upon processing with the program ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The maximum likelihood phylogenetic tree of AFM-1 was constructed by MEGA-X software with 1000 bootstrap replications. The molecular weight and pI value of AFM-1 were predicted using ProtParam2. The putative signal peptide cleavage site of blaAFM-1 was identified by SignalP 5.0. Comparisons of the nucleotide sequences were performed using BLASTN. A plasmid map was generated using GView. Other bioinformatics tools were written using Python and Biopython.
Cloning Experiments
Six ARGs related to antibiotic resistance profiles of A. hydrophila SS332 were chosen for cloning, including blaAFM-1, blaOXA-1, msr(E), mph(E), aac(6ʹ)-Ib10, and aph(3ʹ)-Ia. The primers of the ARGs with the predicted promoter regions were designed and then synthesized by TSINGKE (Beijing, China) (Table S1). For cloning, XbaI (Takara) and BamHI (Takara) restriction endonuclease sites and their protective bases were incorporated into the primers (Table S1, underlined letters). The ARG sequences were PCR amplified from A. hydrophila SS332 genomic DNA with the primers. The PCR products were digested with restriction endonucleases XbaI and BamHI and then ligated into an expression vector (pUCP20 or pUCP24) with T4 DNA ligase (Takara). The recombinant plasmids were transformed into E. coli DH5α, which were then grown on Luria-Bertani agar plates supplemented with ampicillin (100 mg/L) or gentamicin (20 mg/L), and then further verified by colony PCR and sequencing. Antimicrobial susceptibility testing for the transformants was performed using the agar dilution method to verify the function of ARGs. Strains DH5α/pUCP20 and DH5α/pUCP24 were used as controls. For the AFM-1 inhibition assay, the transformants were incubated with 0.05 mmol/L EDTA.
Results
Antibiotic Resistance Phenotypes and Genome Analysis of A. hydrophila SS332
Antimicrobial susceptibility results revealed that of 24 antibiotics tested, A. hydrophila SS332 showed high levels of resistance to 15 antibiotics, especially those with MIC levels at or above 1024 μg/mL, including ampicillin, cefazolin, ceftriaxone, aztreonam, spectinomycin, and roxithromycin. However, SS332 was susceptible to imipenem, meropenem, gentamicin, amikacin, chloramphenicol, florfenicol, levofloxacin, ciprofloxacin and polymyxin B (Table 1).
 |
Table 1 Antibiotic susceptibility profiles of A. hydrophila SS332 and transformants |
To better understand the molecular resistance mechanism of SS332, the complete genome sequence of this strain was determined. The genome of SS332 consists of a chromosome (CP071151) and two circular plasmids designated pSS332-218k (CP071152) and pSS332-5k (CP071153). The chromosome comprises 4,792,137 bp with 60.77% G+C content (Table 2). It contains 4474 coding sequences (CDSs), of which 85.09% are protein coding genes with an average ORF length of 911 bp. A total of 122 tRNAs and 10 rRNA operons were predicted. The genome of pSS332-218k is 218,315 bp with 55.30% G+C content, and a total of 243 CDSs were identified with an average length of 749 bp (Table 2).
 |
Table 2 General features of Aeromonas hydrophila SS332 genome |
In addition, a total of 5 known (100% identity) and 59 putative ARGs (>40% identity) were predicted within the whole genome, of which 52 ARGs (such as blaPER-3, mphA, blaMOX-7, blaOXA-427, aadA16, and mcr-7.1) were located on the chromosome (Table S2), 12 ARGs (including blaAFM-1, blaOXA-1, msr(E), mph(E), aac(6ʹ)-Ib10, and aph(3ʹ)-Ia) were located on the plasmid pSS332-218k (Table S3), and no ARGs were found on plasmid pSS332-5k. Notably, blaAFM-1, a blaNDM-like gene (804 bp), displayed the highest identity (85%) with the known resistance gene blaNDM-6. This gene was first discovered in plasmid pAN70-1 of Alcaligenes faecalis strain AN70 (GenBank: MK757441.1) in the NCBI database, but its functional and genetic characterization has not been performed thus far.
ARGs in pSS332-218k Reduced Antimicrobial Susceptibility
To clarify the role of pSS332-218k in the multiple antibiotic resistance phenotypes of SS332, 6 ARGs carried by pSS332-218k were successfully cloned, including blaAFM-1, blaOXA-1, aac(6ʹ)-Ib10, aph(3ʹ)-Ia, msr(E) and mph(E). The MICs of antibiotics against the recombinant strains are shown in Table 1. DH5α/pUCP24-blaAFM-1 exhibited more than 8-fold higher MICs than DH5α/pUCP24 for ampicillin (16 μg/mL), cefazolin (16 μg/mL), cefoxitin (16 μg/mL), and ceftazidime (8 μg/mL), suggesting that AFM-1 was functional in E. coli DH5α. The expression of OXA-1 in E. coli DH5α conferred increased resistance to ampicillin compared to the control strain. The expression of AAC(6ʹ)-Ib10 and APH(3ʹ)-Ia conferred resistance to kanamycin, with MICs of 128 and 512 μg/mL, respectively. Meanwhile, msr(E) and mph(E) conferred resistance to erythromycin and roxithromycin. These transformants exhibited reduced susceptibility to several tested antibiotics, indicating that 6 ARGs on pSS332-218k could be involved in the antibiotic resistance phenotypes of SS332.
Comparative Genomic Analysis of pSS332-218k
To further investigate the molecular characteristics of pSS332-218k, comparative genomic analysis was conducted. Only one sequence with >50% query coverage (75% coverage) with the pSS332-218k sequence was found by the BLASTN program, which is a complete sequence of plasmid pMCR5-045096 (NZ_CP028567) of A. hydrophila strain WCHAH045096. WCHAH045096 was isolated from sewage in China in 2015. Comparative genomic analysis showed that pSS332-218k and pMCR5_045096 shared a conserved backbone. However, the msr(E), mph(E), blaOXA-1, and blaAFM-1 genes were missing in pMCR5_045096 (Figure 1). Comparative genomic analysis suggested that these ARGs might be acquired by horizontal gene transfer.
Phylogenetic Analysis
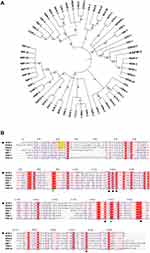
AFM-1, encoded by blaAFM-1, was 267 amino acids in length and displayed the highest level of identity with NDM β–lactamases (84–86%), followed by ElBla2 (55.79%, ABC63608.1) and FIM-1 (42%, AFV91534.1). A phylogenetic tree was created using non-redundant MBLs of subclass B1. Phylogenetic analysis showed that the closest relatives of AFM-1 were NDM-type enzymes (Figure 2A).
Sequence alignment analysis of AFM-1 and representative MBLs of subclass B1 showed that AFM-1 contains conserved zinc-binding residues, namely, H117, H119, D121, H186, C205 and H247 (black spots, Figure 2B).15 The inhibition assay indicated that the enzyme activity of AFM-1 was inhibited by EDTA (Figure S1). The requirement for metal ions (usually Zn2+) is the key feature of the catalytic activity of MBLs.16 The D87 and D189 residues are essential for the structure and stability of NDM-1 by forming hydrogen bonds around Zn2 and Zn1, respectively (green spots, Figure 2B).17 A type II signal peptide cleavage site (yellow box, Figure 2B) was predicted between amino acid residues 22 and 23 and yielded a mature protein of 244 amino acids (theoretical mass, 25,991.23 g/mol; predicted pI, 5.7). AFM-1 has seven amino acid substitutions (V85L, A96S, D127G, A135P, S241N, M242T, and V244A) (yellow box, Figure S2) compared with NDM-1 in structurally conserved sequence fragments (black box, Figure S2) according to the sequence alignment of NDM-β-lactamases (Figure S2).15
Genetic Context of blaAFM-1
Using BLASTN against the GenBank nr database with blaAFM-1 of SS332 as a query, four sequences identical to blaAFM-1 were retrieved, including pAN70-1 (MK757441.1) from Alcaligenes faecalis strain AN70, plasmid pNFYY023-1 (MT011984.1) from Comamonas testosteroni strain NFYY023, Bordetella trematum E202 chromosome (CP049957.1) and Stenotrophomonas maltophilia NCTC10498 chromosome (CP049956.1). To fully elucidate the genetic context of blaAFM-1, comparative genomic analysis was performed on the blaAFM-1-flanking regions of approximately 30 kb in the five blaAFM-1-carrying sequences, including one from this work (Figure 3A).
The blaAFM-1 gene in pSS332-218k was located upstream of a Tn3-type transposon carrying the dfrA5 gene and downstream of an aph(3ʹ)-Ia gene surrounded by two copies of IS26. A conserved fragment (blaAFM-1-bleMBL-trpF-ISCR19-2-msrB-msrA-yfcG-corA) was found around blaAFM-1. Furthermore, the sequences between blaAFM-1 and ISCR19-2 were identical in these five blaAFM-1-carrying sequences, but the sequence between msrB and corA in NCTC10498 shared only 97% identity with that of the other four sequences. MsrA/B encode multidrug efflux pumps that play a positive role in the protection of cellular proteins from oxidative stress damage.18 A similar structure (msrB-msrA-ygcG-corA) has also been found around other β-lactamases, such as blaNDM-1 from Pseudomonas asiatica.19 The conserved fragment containing blaAFM-1 may improve the adaptability of host bacteria to the environment and might aggravate the spread of this gene among different bacteria.
The blaAFM-1 gene in pSS332-218k was associated with three novel ISCR19-like elements or their truncated genes, ISCR19-1, ISCR19-2 and ∆ISCR19-3 (Figure 3B). ISCR19-1 and ISCR19-2 contain intact transposase genes, but ISCR19-3 is interrupted at the 5ʹ end (within the transposase gene). The transposases of these three elements were 1542 bp, 1254 bp and 846 bp in length, respectively, and shared 91.0%, 94.0% and 89.1% nucleotide identity to that of ISCR19, respectively. The oriIS site was identified 245 bp downstream of the stop codon of the transposase gene of ISCR19-1, ISCR19-2 and ISCR19-3. These sequences were conserved in ISCR19-like elements (Figure 3B). The terIS site was identified 135 bp upstream of the beginning of the transposase gene of ISCR19-1 but was absent around the corresponding regions of ISCR19-2 and ∆ISCR19-3.
ISCR19-2 was found in all blaAFM-1-carrying sequences, but ISCR19-1 was not identified in pNFYY023-1, and ∆ISCR19-3 was not found in NCTC10498. Interestingly, ISCR19-1, ISCR19-2 and ∆ISCR19-3 were associated only with blaAFM-1 in GenBank. The results suggested that ISCR19-like elements were involved in the capture and mobilization of blaAFM-1, which was similar to the functions of other ISCR-related β-lactamases, such as blaSPM-1, blaAIM-1 and blaFIM–1.20–22 According to the direction of these ISCR19-like elements, we infer that ISCR19-1 plays a major role in the dissemination of conserved fragments, while ISCR19-2 was initially involved in the capture of msrB-msrA-ygcG-corA regions.
Discussion
In this study, a clinical isolate of A. hydrophila was found to be highly resistant to most tested β-lactam, aminoglycoside, and macrolide antibiotics (Table 1). We observed that DH5α/pUCP24-blaAFM-1 not only induced resistance to ampicillin, cefazolin, cefoxitin, and ceftazidime but also induced low-level resistance to cefotaxime, ceftriaxone, cefoselis, and cefepime, which are 3rd- and 4th-generation cephalosporins. MICs for these cephalosporins were higher for DH5α/pUCP24-blaAFM-1 than for DH5α/pUCP24, with a more than 8-fold increase for cefotaxime and ceftriaxone, a 4-fold increase for cefoselis, and a 2-fold increase for cefepime (Table 1). In addition, whole-genome sequencing analysis revealed that A. hydrophila SS332 possess three chromosomal resistance gene, including extended-spectrum β-lactamases gene blaPER-3, AmpC β-lactamases gene blaMOX-7, and carbapenem-hydrolyzing oxacillinase gene blaOXA-427 (Tables S2). MOX-family and PER-family enzymes hydrolyze extended-spectrum cephalosporins.23,24 OXA-427 reduced susceptibility to carbapenems and conferred resistance to ceftazidime.25 It might be one reason for the high-level resistance of A. hydrophila SS332 to β-lactam antibiotics, including penicillins, monobactams, cephalosporins, and even 3rd- and 4th-generation cephalosporins. However, blaAFM-1 is unable to hydrolyze imipenem and meropenem. We suspect that A. hydrophila SS332 induces low-level resistance to imipenem and meropenem, possibly through chromosomal blaOXA gene or efflux pump genes (Table S2). A previous seven-year surveillance study demonstrated that A. hydrophila (65.6%, 745/1135) was the most predominant species among clinical Aeromonas spp. in Southwest China from 2011 to 2017.26 And this report also mentioned that A. hydrophila has a resistance rates of 30.2% to ceftriaxone; 20% to ceftazidime, cefepime, aztreonam, and ciprofloxacin; and less than 10% to imipenem and meropenem. These findings suggest that the drug resistance of A. hydrophila is an increasingly serious problem in clinical infections. However, the role of plasmids in the antibiotic resistance of A. hydrophila is poorly investigated. pSS332-218k, a multidrug-resistance plasmid from A. hydrophila SS332, carries the antibiotic resistance genes blaAFM-1, blaOXA-1, msr(E), mph(E), aac(6ʹ)-Ib10, and aph(3ʹ)-Ia, which conferred resistance to many of the tested antibiotics when expressed in E. coli, suggesting that these genes are functional (Table S3). Another multidrug-resistance plasmid, pR148, was also obtained from A. hydrophila and had entirely different ARGs, qacH, blaOXA-10, aadA1, sul1, tetA, tetR, and catA2.27 Plasmids are mobile genetic elements that can transfer ARGs to bacterial chromosomes or gain ARGs through horizontal gene transfer. Therefore, determining ARG transmission may be helpful for decelerating the spread of ARGs.
A conserved genomic DNA fragment (blaAFM-1-bleMBL-trpF-ISCR19-2-msrB-msrA-yfcG-corA) containing blaAFM-1 was found in all five different AFM-1-producing strains, two of which were located on chromosomes, and the other three of which were located on plasmids (Figure 3A). These results indicated that the blaAFM-1 gene spread among different strains. Novel ISCR19-like elements were found in the region near to AFM-1 (Figure 3B), suggesting that AFM-1 might be transferred from one strain to another, mediated by ISCR through rolling-circle transposition.28 The acquisition and spread of ARGs is the predominant factor for the escalation of antibiotic resistance.29 ISCR elements are unique mobile elements and can transpose adjacent DNA sequences with a single copy of the element.28 The most worrying feature is that ISCR elements are increasingly associated with genes encoding resistance to extended-spectrum β-lactamases, such as blaPER-3 and blaOXA-18.30,31
Since it was first reported in 2009, NDM-1 has become a great public health concern due to its high carbapenem resistance and global dissemination.32 To date, 27 variants of NDM β-lactamase with 1~5 amino acid substitutions have been reported; in NDM-18, an insertion of five amino acids has been found.33 Among the 28 NDM variants, substitutions of amino acids were identified at 20 different positions. Compared to other NDM variants, AFM-1 has more changes (42 amino acid substitutions) in amino acid residues and shares a similarity of 86% with NDM-1 (Figure 2 and S1). However, the position that plays the critical role in the enzymatic activity of NDM remains unclear. Further studies, such as site-directed mutagenesis, are needed to clarify this issue. Nevertheless, AFM-1 possesses essential residues class that is indispensable for catalytic activity and a substrate-specific residue class that is an alternative for ampicillin, cefotaxime, or imipenem hydrolysis.17 (Figure S2). The functions of essential residues and substrate-specific residues for NDM−1 β-lactamase hydrolysis have been well proven As AFM-1 exhibited greatly reduced resistance to all β-lactam antibiotics, it is possible that the substituted residue(s) are responsible for attenuated β-lactam resistance activity of AFM-1 compared with NDM-1.
Conclusion
In this work, the complete genome sequence of multidrug-resistant A. hydrophila SS332 was determined. Six ARGs (blaAFM-1, msr(E), mph(E), aac(6ʹ)-Ib10, and aph(3ʹ)-Ia) on plasmid pSS332-218k were proven to be functional in E. coli DH5α. Most importantly, a novel blaNDM-like gene, blaAFM-1, was cloned for the first time and found to confer resistance to ampicillin, cefazolin, cefoxitin and ceftazidime. However, AFM-1 exhibited a reduced substrate profile and increased MIC value, which may be caused by amino acid substitutions in structurally conserved sequence fragments. Further analysis of the genetic environment of blaAFM-1 suggested that the risk of spread of blaAFM-1 among different strains, which was mediated by novel ISCR19-like elements.
Ethics Approval and Consent to Participate
Individual patient data were not involved, and only anonymous clinical residual samples were used in this study. This study followed the principles stated in the Declaration of Helsinki.
Acknowledgments
The authors thank all the colleagues and the reviewers who helped with this work. This study was supported by the Natural Science Foundation of Zhejiang Province, China [LQ17H010003, LY19C060002 and LQ17H190001], the National Natural Science Foundation of China [81700011, 81973382 and 81960381], the Science & Technology Project of Wenzhou City, China [Y20170205 and 2019Y0358] and the Science & Technology Project of Inner Mongolia Autonomous Region, China (201802125).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Abdelhamed H, Banes M, Karsi A, et al. Recombinant ATPase of virulent Aeromonas hydrophila protects channel catfish against motile Aeromonas septicemia. Front Immunol. 2019;10:1–7. doi:10.3389/fimmu.2019.01641
2. Xu YG, Chai LH, Shi W, et al. Transcriptome profiling and digital gene expression analysis of the skin of Dybowski’s frog (Rana dybowskii) exposed to Aeromonas hydrophila. Appl Microbiol Biotechnol. 2017;101:5799–5808. doi:10.1007/s00253-017-8385-3
3. Liu JQ, Xie LF, Zhao D, et al. A fatal diarrhoea outbreak in farm-raised Deinagkistrodon acutus in China is newly linked to potentially zoonotic Aeromonas hydrophila. Transbound Emerg Dis. 2019;66:287–298. doi:10.1111/tbed.13020
4. Grim CJ, Kozlova EV, Sha J, et al. Characterization of Aeromonas hydrophila wound pathotypes by comparative genomic and functional analyses of virulence genes. Mbio. 2013;4(2):1–13. doi:10.1128/mBio.00064-13
5. Sedlacek I, Krejci E, Andelova A, et al. Aeromonas hydrophila subsp. dhakensis--a causative agent of gastroenteritis imported into the Czech Republic. Ann Agric Environ Med. 2012;19:409–413.
6. de la Morena ML, Van R, Singh K, et al. Diarrhea associated with Aeromonas species in children in day care centers. J Infect Dis. 1993;168:215–218. doi:10.1093/infdis/168.1.215
7. Chern CH, How CK, Huang LJ. Images in emergency medicine. Necrotizing fasciitis caused by Aeromonas hydrophila. Ann Emerg Med. 2006;48(2):216–225. doi:10.1016/j.annemergmed.2005.12.027
8. Riley PA, Parasakthi N, Liam CK. Development of Aeromonas hydrophila bacteremia in a patient recovering from cholera. Clin Infect Dis. 1996;22:867–868. doi:10.1093/clinids/22.5.867
9. Kali A, Kalaivani R, Charles P, et al. Aeromonas hydrophila meningitis and fulminant sepsis in preterm newborn: a case report and review of literature. Indian J Med Microbiol. 2016;34:544–547. doi:10.4103/0255-0857.195383
10. Fang JS, Chen JB, Chen WJ, et al. Haemolytic-uraemic syndrome in an adult male with Aeromonas hydrophila enterocolitis. Nephrol Dial Transplant. 1999;14:439–440. doi:10.1093/ndt/14.2.439
11. Hughes HY, Conlan SP, Lau AF, et al. Detection and whole-genome sequencing of carbapenemase-producing Aeromonas hydrophila isolates from routine perirectal surveillance culture. J Clin Microbiol. 2016;54(4):1167–1170. doi:10.1128/JCM.03229-15
12. Sekizuka T, Inamine Y, Segawa T, et al. Potential KPC-2 carbapenemase reservoir of environmental Aeromonas hydrophila and Aeromonas caviae isolates from the effluent of an urban wastewater treatment plant in Japan. Environ Microbiol Rep. 2019;11(4):589–597. doi:10.1111/1758-2229.12772
13. Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013;13:1057–1098.
14. Buckner M, Ciusa ML, Piddock L. Strategies to combat antimicrobial resistance: anti-plasmid and plasmid curing. Fems Microbiol Rev. 2018;42(6):781–804. doi:10.1093/femsre/fuy031
15. Garau G, Garcia-Saez I, Bebrone C, et al. Update of the standard numbering scheme for class B beta-lactamases. Antimicrob Agents Ch. 2004;48(7):2347–2349. doi:10.1128/AAC.48.7.2347-2349.2004
16. Jiang XW, Cheng H, Huo YY, et al. Biochemical and genetic characterization of a novel metallo-beta-lactamase from marine bacterium Erythrobacter litoralis HTCC 2594. Sci Rep. 2018;8:1–9. doi:10.1038/s41598-017-17765-5
17. Sun Z, Hu L, Sankaran B, et al. Differential active site requirements for NDM-1 beta-lactamase hydrolysis of carbapenem versus penicillin and cephalosporin antibiotics. Nat Commun. 2018;9:1–14. doi:10.1038/s41467-018-06839-1
18. Soriani FM, Kress MR, Fagundes DGP, et al. Functional characterization of the Aspergillus nidulans methionine sulfoxide reductases (msrA and msrB). Fungal Genet Biol. 2009;46:410–417. doi:10.1016/j.fgb.2009.01.004
19. Tohya M, Tada T, Watanabe S, et al. Emergence of carbapenem-resistant Pseudomonas asiatica producing NDM-1 and VIM-2 metallo-beta-Lactamases in Myanmar. Antimicrob Agents Ch. 2019;63(8):1–8. doi:10.1128/AAC.00475-19
20. Berglund F, Marathe NP, Osterlund T, et al. Identification of 76 novel B1 metallo-beta-lactamases through large-scale screening of genomic and metagenomic data. Microbiome. 2017;5(1):1–12. doi:10.1186/s40168-017-0353-8
21. Pollini S, Maradei S, Pecile P, et al. FIM-1, a new acquired metallo-beta-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy. Antimicrob Agents Ch. 2013;57(1):410–416. doi:10.1128/AAC.01953-12
22. Yong D, Toleman MA, Bell J, et al. Genetic and biochemical characterization of an acquired subgroup B3 metallo-beta-lactamase gene, blaAIM-1, and its unique genetic context in Pseudomonas aeruginosa from Australia. Antimicrob Agents Ch. 2012;56(12):6154–6159. doi:10.1128/AAC.05654-11
23. Ebmeyer S, Kristiansson E, Larsson D. CMY-1/MOX-family AmpC beta-lactamases MOX-1, MOX-2 and MOX-9 were mobilized independently from three Aeromonas species. J Antimicrob Chemoth. 2019;74(5):1202–1206. doi:10.1093/jac/dkz025
24. Li R, Chan EW, Chen S. Characterisation of a chromosomally-encoded extended-spectrum beta-lactamase gene blaPER-3 in Aeromonas caviae of chicken origin. Int J Antimicrob Ag. 2016;47(1):103–105. doi:10.1016/j.ijantimicag.2015.10.018
25. Bogaerts P, Naas T, Saegeman V, et al. OXA-427, a new plasmid-borne carbapenem-hydrolysing class D beta-lactamase in Enterobacteriaceae. J Antimicrob Chemoth. 2017;72(9):2469–2477. doi:10.1093/jac/dkx184
26. Yang S, He T, Sun J, et al. Distinct antimicrobial resistance profiling of clinically important Aeromonas spp. in Southwest China: a seven-year surveillance study. Infect Drug Resist. 2019;12:971–978. doi:10.2147/IDR.S216926
27. Del CC, Hikima J, Jang HB, et al. Comparative sequence analysis of a multidrug-resistant plasmid from Aeromonas hydrophila. Antimicrob Agents Ch. 2013;57(1):120–129. doi:10.1128/AAC.01239-12
28. Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev. 2006;70(2):296–316. doi:10.1128/MMBR.00048-05
29. Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi:10.1038/35012500
30. Wu CJ, Chuang YC, Lee MF, et al. Bacteremia due to extended-spectrum-β-lactamase-producing Aeromonas spp. at a medical center in Southern Taiwan. Antimicrob Agents Ch. 2011;55(12):5813–5818. doi:10.1128/AAC.00634-11
31. Naas T, Namdari F, Bogaerts P, et al. Genetic structure associated with blaOXA-18, encoding a clavulanic acid-inhibited extended-spectrum oxacillinase. Antimicrob Agents Ch. 2008;52(11):3898–3904. doi:10.1128/AAC.00403-08
32. Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Ch. 2009;53(12):5046–5054. doi:10.1128/AAC.00774-09
33. Liu DJ, Piccirilli A, Liu DJ, Li W, Wang Y, Shen J. Deciphering the role of V88L substitution in NDM-24 metallo-β-lactamase. Catalysts. 2019;9:1–11. doi:10.3390/catal9090744
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.