Back to Journals » OncoTargets and Therapy » Volume 13
Molecular Analysis of Oncogenic Mutations in Resected Margins by Next-Generation Sequencing Predicts Relapse in Non-Small Cell Lung Cancer Patients
Authors Wei W, Li X, Song M, Wang C
Received 13 April 2020
Accepted for publication 2 September 2020
Published 25 September 2020 Volume 2020:13 Pages 9525—9531
DOI https://doi.org/10.2147/OTT.S257991
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Federico Perche
Weitian Wei,1– 3 Xingliang Li,4 Mengmeng Song,5 Changchun Wang1– 3
1Institute of Cancer and Basic Medicine (ICBM), Chinese Academy of Sciences, Hangzhou 310022, People’s Republic of China; 2Department of Thoracic Oncology Surgery, Cancer Hospital of University of Chinese Academy of Sciences, Hangzhou 310022, People’s Republic of China; 3Department of Thoracic Oncology Surgery, Zhejiang Cancer Hospital, Hangzhou 310022, People’s Republic of China; 4Department of Thoracic Disease Diagnosis and Treatment Center, Zhejiang Rongjun Hospital, Jiaxing, Zhejiang 314000, People’s Republic of China; 5Geneplus-Beijing Institute, Beijing 102206, People’s Republic of China
Correspondence: Changchun Wang
Department of Medical Oncology, Zhejiang Cancer Hospital, No. 1, East Banshan Road, Gongshu District, Hangzhou 310022, People’s Republic of China
Email [email protected]
Objective: To investigate the genetic mutations in both tumor and marginal tissues in patients with non-small cell lung cancer (NSCLC), and to evaluate the potential prognostic value in patients with margins gene positive.
Methods: Next-generation sequencing (NGS) technique was used to detect genetic mutation in tumor and marginal tissues of the bronchus in 88 patients with NSCLC. Correlation of genetic mutations with pathology, lymph node metastasis, disease-free survival and overall survival was analyzed.
Results: Of the 88 patients, 83 cases (94.3%) had gene mutations in the tumor samples and 12 cases (13.6%) had genetic alterations in their margins. Most of the gene mutations detected were cancer drivers. Six common driver genes between tumor and marginal tissues were identified, including EGFR, TP53, CDKN2A, CTNNB1, BRAF, and NF1. Kaplan–Meier analysis revealed that the median disease-free survival (DFS) was significantly shorter in patients with detectable gene mutations in marginal tissues compared with patients without mutations in margins (30.7 versus 24.4 months, log-rank χ2 = 4.78, P =0.029). Consistently, a shorter median OS was observed in patients harboring gene mutations in margins compared with patients with no mutations in margins (49.1 versus 32.2 months, log-rank χ2 = 3.669, P =0.055).
Conclusion: These findings identify the presence of oncogenic alterations in microscopically negative margins in NSCLC patients associated with elevated risk of relapse and shorter survival time. Thus, examination of microscopically negative margins by NGS represents a valuable approach to predict the clinical outcome of NSCLC patients.
Keywords: lung cancer, gene mutation, surgical margin, next-generation sequencing
Introduction
Lung cancer is the leading cause of cancer-related mortality in the world, with non-small cell lung cancer (NSCLC) accounting for 80–85% of all lung cancer cases.1 Currently, surgical resection is the most effective treatment modality of NSCLC, which is predominately based on clinical staging systems.2 After curative surgery of early NSCLC, the risk of recurrence and prognosis are mainly determined by histopathologic characteristics including histologic type, tumor size, differentiation, and lymph-vascular invasion.
In general, the 5-year survival rate of NSCLC patients with surgery is only about 30–60%, with local recurrence and metastasis being the primary causes of the low survival rate. According to the National Comprehensive Cancer Network, patients with local advanced lung adenocarcinoma are recommended to undergo adjuvant chemotherapy and/or radiotherapy after curative surgery.3 A consistent conclusion whether chemotherapy and/or radiotherapy should be conducted following surgery has not been made on those patients with early stage NSCLC. The majority of patients with stage IA and IB only undergo surgical resection. Although early stage NSCLC cases have a relatively better prognosis, they still experience a 20–30% risk of recurrence after a curative resection.4 Unfortunately, it still remains unclear about the molecular mechanisms of recurrence following curative surgery in patients with early stage NSCLC. Identification of patients with high risk of recurrence may contribute to guide personalized treatment strategy for the individual patient, in particular for those who may benefit from adjuvant chemotherapy and/or radiotherapy.
Accumulating studies reveal that microscopically normal-appearing tissues adjacent to cancers harbor genetic, epigenetic or transcriptional alterations in a variety of cancer types.5,6 TP53 gene mutations, methylation of specific genes and gene profiling in the non-malignant tissues are considered as potential biomarkers for early detection of lung cancer.7–9 It is suggested that the malignant molecular changes appear long before the morphological changes, serving as “molecular margins” in the assessment of surgical margins of lung cancer.
With the development of next-generation sequencing (NGS) technology, it is feasible to identify oncogenic alterations that may be missed by conventional pathological techniques.10 In the present study, we depicted the landscape of gene mutations in both tumor and marginal tissue in patients with NSCLC. Furthermore, we explored the diagnostic potential of genetic alterations in marginal tissues in NSCLC.
Methods
Consecutive patients who underwent complete resection with systematic lymph node dissection were retrospectively analyzed between 2013 and 2014. All patients had pathologic R0 resection and margin distance of 1 to 2 cm. The free resection margin was defined as the nearest distance between the tumor and the resection line. No patients received chemotherapy or radiotherapy before the operation. Microscopically negative margin tissue was used for gene detection. All of the pathology results were evaluated by two independent pathologists. All the specimens of this project were from the biological sample bank of Zhejiang Cancer Hospital. All patients signed the informed consent before the specimens were put into storage. The study was approved by the institutional ethics committee of Zhejiang Cancer Hospital (approval no. IRB-2018-24).
Sample Processing and DNA Extraction
Isolation of intact total DNA from FFPE tumor and marginal tissue samples was carried out using Max-well® RSC DNA FFPE Kit (Promega, Madison, WI, USA). DNA quantification was measured using Qubit fluorometer with Qubit dsDNA HS (High Sensitivity) Assay Kit (Invitrogen, Carlsbad, CA, USA). The size distribution analysis of DNA fragment used Agilent 2100 BioAnalyzer and the DNA HS kit (Agilent Technologies, Santa Clara, CA, USA).
Library Construction and Target Enrichment
The genomic DNA extracted from FFPE specimens was sheared to 300-bp fragments with a Covaris S2 ultrasonicator (Covaris, Woburn, MA, USA) before library construction. A volume of 100–500 ng DNA from the FFPE specimen was used for library construction. Fragmented libraries for next-generation sequencing (NGS) were constructed using the KAPA Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA).
We designed the probe to explore genetic properties of NSCLC based on ~230-kb genomic regions of 59 genes frequently mutated in NSCLC and other common solid tumors. Enrichment of target was performed with a custom SeqCap EZ Library (Roche NimbleGen, Madison, WI, USA). DNA hybridization capture was carried out according to the manufacturer’s protocol. Following hybrid selection, the captured DNA fragments were amplified and then pooled to generate several multiplex libraries.
NGS Sequencing
Paired-end 2 × 75 reads sequencing was carried out on an Illumina HiSeq 3000 instrument according to the manufacturer’s recommendations using TrueSeq PE Cluster Generation Kit v3 and the TrueSeq SBS Kit v3 (Illumina, San Diego, CA, USA).
Data Analysis
After filtering low quality data and removing adaptor sequences, clean reads were mapped to the reference human genome (hg19) and aligned using BWA-mem. Single nucleotide variants (SNVs) and somatic small insertions and deletions (InDel) calling used Mutect2 software. VAF (also called mutant allele burden) was defined as a read count supporting the mutant base divided by the total read count at that position. All final candidate variants were manually verified with the integrative genomics viewer browser. Statistical analyses were performed using GraphPad Prism (7.04).
Statistical Analysis
Disease-free survival (DFS), was defined as the time from confirmed pathology to recurrence or metastasis. OS was calculated as the period from the date of surgery to the date of any cause of death or the last follow-up time. DFS and OS were analyzed using the Kaplan–Meier method and compared between different groups using the Log rank test. Unpaired Wilcoxon signed-rank test was used for continuous variable comparison, and two-sided Fisher’s exact test was used to compare categorical data, as appropriate. P < 0.05 was considered statistically significant. Statistical analyses were performed using GraphPad Prism (7.04). The median follow-up time was 44.5 months (range 7–65).
Results
Clinicopathologic Characteristics of Present Study
Eighty-eight patients were identified in present study. The median age was 60 years old (range, 41–79 years); 63 were male and 25 female. Thirty-three subjects were ever-smokers or smokers, whereas 55 were non-smokers. Fifty-six were diagnosed with adenocarcinoma and 32 with squamous cell carcinoma. According to the 8th edition of the tumor, node, metastasis classification system staging system, 43 patients (14.9%) were classified as having 65 of stage I and II, 23 of IIIA disease. All of the 88 cases were invasive adenocarcinoma. Of those, 25, 18, 16, 14, 13, and 2 were papillary predominant, acinar predominant, micropapillary predominant, solid predominant, lepidic predominant subtypes, and variants of invasive adenocarcinoma, respectively. No subtype difference was found in patients with and without tissue margin positive.
The clinicopathologic characteristics are listed in Table 1.
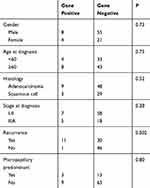 |
Table 1 Clinicopathologic Features Comparison Between Patients with Margins Gene Positive and Negative |
Landscape of NSCLC and Marginal Tissue Gene Mutations
The number of mutations ranged from 1 to 24 in 83 tumor cases (83/88, 94.3%). TP53 was the most common mutation identified in the tumor tissue (Figure 1A), accounting for 62.6% (52/83) of patients; EGFR was the second most common, accounting for 48.1% (40/83).
Genes mutations were identified in 12 marginal tissue cases (12/88, 13.6%) (Figure 1B). Mutation genes identified in marginal tissues include TP53, EGFR, FGFR1, BRAF, NF1, CDKN2A, CTNNB1, and PMS2. FGFR1 (4/12, 33.3%), BRAF (3/12, 25%), EGFR (3/12, 25%) were the most frequent in marginal tissues. All the genes were lung cancer driver genes, besides FGFR1 and PMS2.
Between the tumor and marginal tissues of 12 patients, lung cancer driver genes were identified in tumor tissues of 11 patients and in marginal tissues of 7 patients. The common driver genes between tumor and marginal tissues of 12 patients were EGFR, TP53, CDKN2A, CTNNB1, BRAF, and NF1 (Figure 1C).
Correlation Between Gene Mutations in Marginal Tissues and Clinical Features
We compared mutation profile (mutation number and minor allele frequency) with different clinical features. There were no statistically significant differences between mutation profiles of clinical features groups (Figure 2).
Furthermore, genetic variables (mutation status and driver gene status) were compared within groups of patients with different clinical features, and also no difference was found between different groups (Figure 3).
Association of Gene Mutations in Marginal Tissues with DFS and OS in Lung Cancer
Among the 88 patients, gene mutations were identified in 12 marginal tissue cases (12/88, 13.6%) and were absent in 76 tumor cases (76/88, 86.4%). Median DFS was significantly shorter in patients with detectable gene mutations in marginal tissues compared with those patients without gene mutations in their marginal tissues (30.7 versus 24.4 months, χ2 = 4.78, P =0.029) (Figure 4). Consistently, an obviously shorter median OS was observed in patients with gene mutations in marginal tissues compared with those without mutations in their margins (49.1 versus 32.2 months, P =0.055) (Figure 5). Collectively, these findings reveal that driver genes in the marginal tissues may be responsible for patients with shorter DFS and OS.
 |
Figure 4 Kaplan–Meier survival curves of DFS in lung cancer patient with margins gene positive (n=12) and negative (n=76). |
 |
Figure 5 Kaplan–Meier survival curves of OS in lung cancer patient with margins gene positive (n=12) and negative (n=76). |
Discussion
Local recurrence or metastasis is mainly responsible for the treatment failure and relapse following initial surgical resection.11 In addition, the molecular status of the “normal”-appearing lung tissue in the surgical margins and its clinical significance has rarely been investigated in NSCLC. In the present study, the gene profiles in both tumor tissues and marginal tissues in lung cancer were evaluated using NGS technology. A total of 12 marginal tissue cases harbored gene alterations, with a majority of them being driver genes for lung cancer. Moreover, our results showed that patients harboring gene mutations in their marginal tissues had shorter DFS and OS, suggesting that these gene mutations may be responsible for the recurrence of this subtype of patients with NSCLC.
Although the early stage NSCLC patients are considered to have better prognosis, with 5-year survival rates ranging from 50% to 70%, nearly 30–50% of them will relapse.12,13 To improve the prognosis of lung cancer, it is important to secure cancer-free surgical margin in curative surgical resection of tumors. The conventional approach to validate a clean surgical resection is based on pathological characteristics on margins of normal tissue.13 However, the microscopic residual (R1) margin rate was 3–7% after curative surgery of lung cancer. The overall survival rate declines from 62% to 37% in the early stage patients with R1 margins.14 Moreover, local recurrence following pulmonary resection occurs in approximately 20% of patients even without microscopic or macroscopic cancer residuals.5
Many researchers have explored many approaches to predict the clinical outcome of early stage patients, including detection of genetic alterations, epigenetic modifications, and gene-expression profiles.15–17 Only a few studies have explored the molecular status of histologically normal-appearing surgical margins.18–20 Masasyesva et al analyzed the molecular margins of sublobar resections of lung cancer and identified that K-RAS mutation at codon 12 was associated with local recurrence of NSCLC.18 Another study by Lv et al found that oncogenic mutations were identified in 87.5% bronchial margins of early relapse patients, suggesting that genetic alterations in the margins may be risk factors of recurrence in lung cancer patients at early stages.19 In our study, we found that oncogene mutations occurred in 94.3% and 13.6% of tumor cases and marginal tissues using NGS, respectively. Most of the gene mutations detected were cancer drivers. Moreover, six common driver genes between tumor and marginal tissues were identified, including EGFR, TP53, and CDKN2A. In addition, we analyzed the correlation between gene mutations in marginal tissues and clinical features and found that advanced-stage patients had a higher percentage of driver gene and a higher percentage of mutated samples than patients with other stages. In addition, we observed shorter median DFS and OS in patients with gene mutations in marginal tissues compared with those with no mutations in their margins, suggesting that driver genes in the margins may contribute to the recurrence and relapse of these patients.
There were several limitations in this study. First, only 88 patients were enrolled and 12 with margin positive. Thus, more patients are needed to validate these findings. Second, this study focused on genetic mutations in the samples, and no liquid biopsy samples were obtained. There is intratumoral heterogeneity in tissue biopsy; however, a systematic review by Stepan et al showed that targeted NGS in plasma shows inferior performance in detecting mutations compared to tissue biopsy in advanced NSCLC patients.21 Last, the mutated genes in the clear tumor margins may be due to infiltrating tumor cells or precancerous cells which harbor the mutations; however, we could not clear the possibilities in present study.
In conclusion, our study reveals that oncogenic alterations are detectable in microscopically negative margins in NSCLC patients, with elevated risk of relapse and shorter survival time. These findings demonstrate that examination of microscopically negative margins by NGS represents a novel “molecular margin”, which is valuable for predicting outcome of NSCLC patients.
Acknowledgments
This study was supported by the Zhejiang Provincial Natural Science - Mathematical Medicine Association Joint Fund (LSY19H160001) and the Medical and Health of Zhejiang Province Scientific Research Project (2013KYA031).
Disclosure
Mengmeng Song is an employee of Geneplus- Beijing Institute. The authors report no other potential conflicts of interest for this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;60(5):277–300.
2. Lou F, Huang J, Sima CS, et al. Patterns of recurrence and second primary lung cancer in early-stage lung cancer survivors followed with routine computed tomography surveillance. J Thorac Cardiovasc Surg. 2013;145(1):75–82. doi:10.1016/j.jtcvs.2012.09.030
3. Osarogiagbon RU, Ray MA, Faris NR, et al. Prognostic value of national comprehensive cancer network lung cancer resection quality criteria. Ann Thorac Surg. 2017;103(5):1557–1565. doi:10.1016/j.athoracsur.2017.01.098
4. Saisho S, Yasuda K, Maeda A, et al. Post-recurrence survival of patients with non-small-cell lung cancer after curative resection with or without induction/adjuvant chemotherapy. Interact Cardiovasc Thorac Surg. 2013;16(2):166–172. doi:10.1093/icvts/ivs450
5. Sawabata N, Maeda H, Matsumura A, Ohta M, Okumura M. Clinical implications of the margin cytology findings and margin/tumor size ratio in patients who underwent pulmonary excision for peripheral non-small cell lung cancer. Surg Today. 2012;42:238–244. doi:10.1007/s00595-011-0031-6
6. Dakubo GD, Jakupciak JP, Birch-Machin MA, Parr RL. Clinical implications and utility of field cancerization. Cancer Cell Int. 2007;7:2. doi:10.1186/1475-2867-7-2
7. Franklin WA, Gazdar AF, Haney J, et al. Widely dispersed p53 mutation in respiratory epithelium. a novel mechanism for field carcinogenesis. J Clin Invest. 1997;100:2133–2137. doi:10.1172/JCI119748
8. Guo M, House MG, Hooker C, et al. Promoter hypermethylation of resected bronchial margins: a field defect of changes? Clin Cancer Res. 2004;10:5131–5136. doi:10.1158/1078-0432.CCR-03-0763
9. Spira A, Beane JE, Shah V, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi:10.1038/nm1556
10. Wang K, Stephens P, Yelensky R, et al. Frequency of actionable genomic alterations in early-stage lung adenocarcinoma (LA) detected by next-generation sequencing (NGS). J Clin Oncol. 2012.
11. Weinberg RA, Chaffer CL. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi:10.1126/science.1203543
12. Choi PJ, Jeong SS, Yoon SS. Prediction and prognostic factors of post-recurrence survival in recurred patients with early-stage NSCLC who underwent complete resection. J Thorac Dis. 2016;8(1):152.
13. Uramoto H, Tanaka F. Recurrence after surgery in patients with NSCLC. Transl Lung Cancer Res. 2014;3(4):242–249.
14. Hancock JG, Rosen JE, Antonicelli A, et al. Impact of adjuvant treatment for microscopic residual disease after non-small cell lung cancer surgery. Ann Thorac Surg. 2015;99(2):406–413. doi:10.1016/j.athoracsur.2014.09.033
15. Jacek JPD, Ewa JPD, Witold RPD, et al. P53 and K‐ras mutations are frequent events in microscopically negative surgical margins from patients with nonsmall cell lung carcinoma. Cancer. 2004;29(1):182–183.
16. Meaburn E, Schulz R. Next generation sequencing in epigenetics: insights and challenges. Semin Cell Dev Biol. 2012;23(2):192–199. doi:10.1016/j.semcdb.2011.10.010
17. Navin N, Krasnitz A, Rodgers L, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20(1):68–80. doi:10.1101/gr.099622.109
18. Masasyesva BG, Tong BC, Brock MV, et al. Molecular margin analysis predicts local recurrence after sublobar resection of lung cancer. Int J Cancer. 2005;113(6):1022–1025. doi:10.1002/ijc.20683
19. Lv T, Zou J, Liu H, et al. Detection of oncogenic mutations in resected bronchial margins by next-generation sequencing indicates early relapse in stage IA lung adenocarcinoma patients. Oncotarget. 2017;8(25):40643–40653. doi:10.18632/oncotarget.16539
20. Cao B, Feng L, Lu D, et al. Prognostic value of molecular events from negative surgical margin of non-small-cell lung cancer. Oncotarget. 2016;8(32):53642.
21. Stepan ME, Georgia ΙG, Ilias PN, et al. Comparison of liquid-based to tissue-based biopsy analysis by targeted next generation sequencing in advanced non-small cell lung cancer: a comprehensive systematic review. J Cancer Res Clin Oncol. 2020;146(8).
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



