Back to Journals » Infection and Drug Resistance » Volume 14
Molecular Analysis and Antimicrobial Resistance Pattern of Tigecycline-Non-Susceptible K. pneumoniae Isolated from a Tertiary Care Hospital of East Asia
Authors Hu N, Wang D, Lin Y, Zou J, Liu Y, Xiong Z, Guo J, Zeng L , Li J
Received 16 August 2021
Accepted for publication 23 September 2021
Published 7 October 2021 Volume 2021:14 Pages 4147—4155
DOI https://doi.org/10.2147/IDR.S334098
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Niya Hu,1,* Dongjiang Wang,2,* Yiqing Lin,1 Jun Zou,3 Yanling Liu,1 Zhigang Xiong,3 Jian Guo,2 Lingbing Zeng,1 Junming Li1
1Department of Clinical Laboratory, The First Affiliated Hospital of Nanchang University, Nanchang, People’s Republic of China; 2Department of Laboratory Medicine, Shanghai East Hospital, Tongji University, School of Medicine, Shanghai, People’s Republic of China; 3Department of Orthopedics, Jiangxi Provincial Children’s Hospital, Nanchang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lingbing Zeng; Junming Li Email [email protected]; [email protected]
Introduction: Tigecycline is one of the last resorts for carbapenem-resistant K. pneumoniae (CRKP) infections. Indeed, tigecycline-non-susceptible K. pneumoniae (TNSKP) strains are increasingly treated with the use of tigecycline. In this study, we attempted to better understand their epidemiological trends and characteristics. K. pneumoniae were collected from 2017 to 2020 at the First Affiliated Hospital of Nanchang University.
Methods: Thirty-four TNSKP strains were selected during the study period, all of which were analyzed using antimicrobial susceptibility testing, multilocus sequence typing (MLST), and pulsed-field gel electrophoresis (PFGE). PCR and DNA sequencing were performed for the detection of β-lactamase genes and carbapenemase genes, and the mutation analysis of tet(A), tet(X), tet(L), tet(M), rpsJ, ramR, and oqxR, which are related to tigecycline resistance. Virulence gene and capsular genotype testing were conducted to identify whether the TNSKP strains were hypervirulent Klebsiella pneumoniae.
Results: An epidemiology analysis showed that Klebsiella pneumoniae carbapenemase-2 (KPC-2) was the predominant carbapenemase in tigecycline non-susceptible carbapenem-resistant K. pneumoniae (TNSCRKP) (96.7%), and the dominant clone type was ST11-K14K64 (82.4%). Among them, 55.9% (19/34) of strains were from each department of ICU, particularly EICU and neurosurgery ICU. In order to further understand the molecular mechanisms of the TNSKP, a polymerase chain reaction of the resistant determinants was carried out. The results detected many tigecycline-resistant genes, such as tet(A) (97.1%), tet(X) (17.6%), rpsJ (97.1%), and ramR (8.8%).
Conclusion: As the results of this study reveal, we should take effective measures to control the increase in TNSKP.
Keywords: carbapenem-resistant, tigecycline-non-susceptible, Klebsiella pneumoniae
Introduction
Klebsiella pneumoniae belongs to the Enterobacteriaceae family and is an opportunistic pathogen that can transfer to multi-drug resistant strains and can cause various infections, including bacteremia, pneumonia, liver abscess, and urinary tract infection.1,2 At present, the spread of multi-drug resistant K. pneumoniae strains is a worldwide problem, and especially the carbapenem-resistant K. pneumoniae (CRKP); their infections are usually associated with high mortality.3
Tigecycline is a novel type of broad-spectrum glycyl-tetracycline antibiotics administered after minocycline development. It has a very wide range of activities against most Gram-positive bacteria, Gram-negative bacteria, and anaerobic bacteria, especially for multi-drug resistant strains.4,5 Tigecycline is one of the few available options for the treatment of carbapenem-resistant bacterial infections.6 Thus, tigecycline is considered to be the last line of defense for the treatment of this kind of bacterial infection. However, tigecycline-insensitive strains of K. pneumoniae are increasingly being reported, with some reports showing emergence in Taiwan,7 Northern China,8 Korea,9 Austria,10 etc. According to the China Antibiotic Resistance Surveillance System for antibiotic resistance, the tigecycline-non-susceptible K. pneumoniae (TNSKP) was up to 8.5% in 2019 (http://www.carss.cn/); thus, we should pay more attention to them in clinical infections.
In order to explore whether tigecycline-non-susceptible K. pneumoniae are also present in the First Affiliated Hospital of Nanchang University, and then to better understand their epidemiological trends and characteristics, we analyzed the K. pneumoniae in the hospital from 2017 to 2020, and screened for tigecycline-non-susceptible K. pneumoniae in order to perform further analysis. We aimed to provide more advice that may be useful for controlling the infection rates in the hospital.
Materials and Methods
Bacterial Strains and Data Collection
In this study, we collected 5729 K. pneumoniae non-duplicate strains from the First Affiliated Hospital of Nanchang University from 2017 to 2020. Cases of infection with tigecycline-non-susceptible K. pneumoniae strains were detected at different wards, and strains were isolated from different types of clinical specimens. Detailed clinical data were obtained from electronic medical records and informed consent was obtained from all patients included in the study. A concise flow chart is shown in Figure 1. All clinical isolates were identified using MALDI-TOF MS (Bruker, Sancordon Inc., Bremen, Germany). K. pneumoniae ATCC 13883 was used as a quality control strain for antimicrobial susceptibility testing. The Salmonella enterica serotype Braenderup strain (H9812) was used as a reference standard of pulsed-field gel electrophoresis (PFGE).
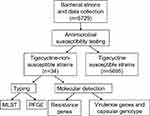 |
Figure 1 A concise flow chart about this study. |
Antimicrobial Susceptibility Testing
The MICs of amoxicillin-clavulanic acid, piperacillin-tazobactam, ceftazidime, ceftriaxone, ertapenem, imipenem, meropenem, amikacin, ciprofloxacin, and tigecycline were determined using the broth microdilution methodology. Except for tigecycline, the sensitivity of other antibacterial agents was determined with reference to the CLSI M100 guidelines (2020). The interpretive criteria for tigecycline was according to US Food and Drug Administration (FDA) standards.
Molecular Detection of Resistance Genes
We carried out polymerase chain reaction detection of β-lactamase genes, including blaTEM, blaSHV, blaCTX-M, and blaLAP, and carbapenemase genes, including blaVIM, blaKPC, blaIMP, blaNDM, and blaOXA-48. Potential tigecycline-resistance determinants containing tet(A), tet(X), tet(L), tet(M), rpsJ, ramR, and oqxR11–13 were also amplified in the tigecycline-non-susceptible strains and sequenced in positive strains; then, rpsJ, ramR, and oqxR were compared with the wild-type reference strain K. pneumoniae MGH78578 (GenBank accession number CP000647) as previously described.11 The primers used to amplify the coding sequence of each gene are listed in Table S1.
Molecular Detection of Virulence Genes and Capsular Genotype
It will be more difficult to treat clinically strains that express both virulence genes and are tigecycline non-susceptible. Thus, we detected virulence genes, including rmpA, aero, uge, ironB, mrkD, and kpn, and we used the wzi capsule gene to determine the capsule genotype (K-type) in the tigecycline non-susceptible strains.
Multilocus Sequence Typing (MLST) and Pulsed-Field Gel Electrophoresis (PFGE) for Tigecycline-Non-Susceptible Strains
According to the protocol described on the MLST Klebsiella pneumoniae website (https://bigsdb.pasteur.fr/klebsiella/primers_used.html), all tigecycline non-susceptible strains were tested with seven housekeeping genes (rpoB, gapA, mdh, pgi, phoE, infB, and tonB14). The results of MLST were analyzed using the international Klebsiella pneumoniae MLST database established at the Pasteur Institute in France in 2005.14 Allele sequence and sequence type (ST) are verified on https://bigsdb.pasteur.fr/klebsiella/. PFGE was performed as previously described,15 the screened tigecycline non-susceptible K. pneumoniae strains were picked from fresh pure colonies into the buffer, and each of them was placed into small plastic containers. The DNA was digested by the restriction enzyme XbaI (TakaRa, Japan) 10U for 4 h at 37 °C. Electrophoresis was conducted using 0.5 × TBE buffer at 14 °C for 18 h, and pulse time was 2–64 s. Salmonella enterica serotype Braenderup strain (H9812) was used as a classical molecular weight standard. We finally used Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium) to analyze homogeneity. Clinical isolates with more than 80% homology were defined as the same PFGE cluster.
Results
Clinical Characteristics of Patients Infected with TNSKP
In this study, a total of 5729 K. pneumoniae strains were collected, there were 1282, 1397, 1564, and 1486 strains in 2017–2020, respectively, and of them 1851 (32.3%, 1851/5729) strains were CRKP. The K. pneumoniae strains showed high susceptibility to TGC, but we also found 34 patients infected with TNSKP; the demographic and clinical characteristics of the 34 strains are shown in Figure 2. Among the 34 isolates, the number of specimens from sputum was the largest, accounting for 64.7% (22/34), and the number of specimens from urine 17.6% (6/34) ranked second, followed by blood 5.9% (2/34), ascites 5.9% (2/34), catheter tip 2.9% (1/34), and wound fluid 2.9% (1/34). The TNSKP strain was not found in 2017, but the TNSKP rates rose during the following years, by 0.1% (2/1397), 0.6% (9/1564), and 1.5% (23/1486) in 2018–2020, respectively. Analysis of the departmental distribution of the patients revealed that 55.9% (19/34) of the patients with non-susceptible strains were from the intensive care unit (ICU), particularly from EICU 14.7% (5/34) and neurosurgery ICU 14.7% (5/34). In 2018, both of the TNSKP strains were from neurosurgery ICU. In addition to this, common departments also had more TNSKP strains, such as the rehabilitation department 11.8% (4/34) and neurosurgery 11.8% (4/34).
Antibiograms
The antibiotic resistance results of TNSKP strains to 10 antibiotics are shown in Table 1. We can see that 34 strains showed low susceptibility to all of the 10 antibiotics, including piperacillin/tazobactam (11.8%), ceftazidime (2.9%), ceftriaxone (2.9%), ertapenem (11.8%), imipenem (14.7%), meropenem (14.7%), amikacin (17.6%), and ciprofloxacin (5.9%); acid susceptibility rates to amoxicillin/clavulanic were zero. Among the 34 tested TNSKP strains, 30 (30/34, 88.2%) were also CRKP strains. According to the interpretive criteria for tigecycline (≤2 μg/mL, susceptible; 4 μg/mL, intermediate; ≥8 μg/mL, resistant), there were three (3/34, 8.8%) intermediate strains; the others were resistant.
 |
Table 1 Antibiotic Susceptibilities of TNSKP Strains (n=34) |
Analysis of Resistance Determinants in TNSKP Strains
To investigate the resistance mechanism of TNSKP, potential tigecycline resistance determinants were identified in 34 strains by PCR and sequencing. The genes tet(A) and rpsJ were detected in most of the TNSKP strains except for strain No. 44; tet(X) was found in six strains (17.6%), whereas tet(L) and tet(M) were not detected in all of the TNSKP strains. Three strains (8.8%) were detected with ramR expression; similarly, oqxR was found in three (8.8%) isolates. One strain with S29L substitutions in ramR and another two strains with a termination codon appeared in advance. Meanwhile, mutations in rpsJ and oqxR were not identified in this study (Table 2).
 |
Table 2 Resistance Determinants and Virulence Genotype Were Examined in the Present Study |
A total of five carbapenemases were detected in this study; 96.7% (29/30) of Tigecycline non-susceptible carbapenem-resistant K. pneumoniae (TNSCRKP) were blakpc positive (Table 2); and blaVIM, blaIMP, blaNDM, and blaOXA-48 were not detected in any of the strains. Among the 34 strains, it was detected that 29 strains carried blaSHV-12, 2 strains carried both blaSHV-158 and blaSHV-187, 30 strains carried blaTEM-1, and 1 strain carried blaTEM-235. A total of 31 strains carried blaCTX-M-65, 1 strain carried both blaCTX-M-3 and blaCTX-M-14, and 31 strains carried blaLAP-2. For strain No. 11 only, no carbapenemases were detected while in CRKP.
Virulence Analysis
All strains in the study carried the mrkD gene, followed by kpn (97.1%, 33/34) and uge (91.2%, 31/34). Only one strain carried the rmpA gene, while aero and iroN were not detected among any of the strains. Two or more genes coexisted in these isolates; the combination of uge, mrkD, and kpn was the most common prototype observed, and there were 29 isolates of this type. Three isolates were a combination of mrkD and kpn, while one combination of mrkD and uge was found. There was only one strain, No. 25, that was a combination of rmpA, uge, mrkD, and kpn (Table 2).
Isolates Typing and PFGE
In total, five different multilocus sequence types were detected; ST11 was the most prevalent type (85.3%, 29/34), followed by ST37 (5.9%, 2/34), and only one strain each of ST3627, ST2593, and ST2601 (Figure 2).
The wzi capsular gene was sequenced to assign a capsular type (K-type) to each isolate. In this study, two K-types were identified. A total of 88.2% (30/34) of the isolates were typed as K14.K64, while 8.8% (3/34) were typed as K15.K17.K50.K51.K52. One strain, No. 63, was not linked to any K-type (Figure 2).
According to the results of PFGE, the similarity of the strains ranged between 60% and 100%. Among them, the 34 strains were divided into six different PFGE clusters (clusters A to F) at an 82% similarity cut-off (Figure 2). Obviously, the dominant strain species was cluster A, which contained a total of 28 strains (82.4%, 28/34), followed by cluster D (5.9%, 2/34); each of the remaining four cluster (B/C/E/F) contained only one strain; 29 ST11 isolates mainly belonged to cluster A.
Discussion
The prevalence of carbapenem-resistant K. pneumoniae has been increasing globally, making antimicrobial treatment difficult and leading to higher disease-related mortality.16,17 In this study, we collected 5729 K. pneumoniae strains from our hospital over four years; carbapenem resistance rates in K. pneumoniae reached 32.3%, which is higher than the average level in Jiangxi province according to the China Antibiotic Resistance Surveillance System in 2019 (http://www.carss.cn/). This may be related to the large number of severe patients in our hospital, and because of long hospital stays, most of the TNSKP strains were from the intensive care unit. With the increasing administration of tigecycline in clinical settings, tigecycline-non-susceptible strains will become increasingly prominent. The TNSKP rates were below 8.5% at present in our hospital, but we should pay more attention to this issue, in light of the rising trend detected over our four-year study period. Meanwhile, we found that most of the TNSKP strains were also resistant to carbapenems, which is consistent with other studies;18 hence, the antimicrobial susceptibility testing confirmed low susceptibility to all of the 10 antibiotics. According to the antibiotic resistance results of tigecycline, there were three intermediate strains (strains No. 38, 44, and 53), and among them two strains with mutations in ramR (strains No. 44 and 53), and two strains were detected with oqxR expression (strains No. 38 and 44). To our knowledge, the AcrAB-TolC efflux pump associated with the inactivation of the repressor ramR caused by mutations is a major cause of tigecycline resistance.19 In addition to this, the OqxAB efflux pump is downregulated by the repressor oqxR, which was also associated with tigecycline resistance.20 Thus, our results are in accordance with the above. Except for efflux-mediated resistance mechanisms, there were other resistance mechanisms such as Tet proteins, including mutated tet(A), tet(L), tet(X), and tet(M), and the rpsJ gene, which was associated with an alteration in the tigecycline target site of the ribosomal protein S10.12 In this study, 31/34 (91.2%) strains were tigecycline-resistant, and all of them were detected as tet(A) and rpsJ gene-positive, although mutations were not identified. We suspect that they may be the main cause of the resistance mechanisms among these strains, but further studies are needed.
At present, common carbapenemase genes, including blaVIM, blaKPC, blaIMP, blaNDM, and blaOXA-48, as well as KPC-type enzymes, and in particular KPC-2, are the most common carbapenemases in China.21,22 As our data show, 96.7% (29/30) of Tigecycline non-susceptible carbapenem-resistant K. pneumoniae were detected with blaKPC-2, which is consistent with previous reports. Only for strain No. 11 was no carbapenemase detected in this study. Strain No. 11 was ertapenem-resistant but imipenem- and meropenem-susceptible, and showed blaSHV-158, blaCTX-M-65, and blaLAP-2 beta-lactamases resistance; previous studies showed that the mechanisms of resistance in this form would be predominantly mediated by non-carbapenemase mechanisms, including deficiencies in the expression of porins Omp35 and Omp3623 or the production of particular beta-lactamases.24
Among the TNSKP isolates, we detected the virulence-related genes mrkD, kpn, and uge with a positive rate more than 90%; these genes are associated with adhesins and lipopolysaccharides of K. pneumoniae.25 Only one strain was positive for the rmpA gene, which is related to the high viscosity of K. pneumoniae;26 aero and iroN, which are associated with iron carriers, were not detected in this study. In addition, in this experiment we determined capsular types by sequencing the wzi gene, and they were mainly the K14.K64 type, whereas previous studies reported the K47 type as dominant.27 In this study, we did not find virulent capsular types, such as K1, K2, K5, K16, K20, K54, and K57, and thus these TNSKP isolates carried high resistance but low virulence.
Molecular typing of our strains showed the dominant sequence type of the TNSKP isolates to be ST11, which is in accordance with reports that ST11 is the most common type of CRKP associated with the spread of blaKPC-2 in China.28,29 Other STs, such as ST37, ST3627, ST2593, and ST2601, were also detected in 34 TNSKP strains, which was different from previous reports in Henan.30 Our study is the first to report of ST3627, ST2593, and ST2601 K. pneumoniae with tigecycline-non-susceptible caused by clinical infection. PFGE results showed that the most prevalent clone type ST11-K14K64 (28/34, 82.4%) belongs to cluster A. In cluster A, the ST11-K14K64 isolates were mainly found in the ICU, neurosurgery ICU, and EICU, which indicated that there may have been a prevalence of ST11-K14K64 TNSKP in the ICU of our hospital from 2018 to 2020.
Conclusion
The rate of carbapenem-resistant K. pneumoniae in our hospital was higher than the average level in Jiangxi province, the tigecycline-non-susceptible K. pneumoniae was often found in carbapenem-resistant strains, and they were usually isolated from the intensive care unit. The ST11-K14K64 clone type carrying the blaKPC-2 was primarily detected in this study, and the tet(A) and rpsJ genes may contribute to the tigecycline-non-susceptible phenotype in the 34 TNSKP strains, which is worthy of further research. In all, we should take effective measures, such as active surveillance and effective segregation to prevent CRKP, thus reducing the emergence of TNSKP.
Abbreviations
CRKP, carbapenem-resistant K. pneumoniae; TNSKP, tigecycline-non-susceptible K. pneumoniae; TNSCRKP, tigecycline non-susceptible carbapenem-resistant K. pneumoniae; PFGE, pulsed-field gel electrophoresis; CLSI, the Clinical and Laboratory Standards Institute; FDA, Food and Drug Administration; MLST, multilocus sequence typing; ST, sequence type; ICU, intensive care unit.
Data Sharing Statement
The data used and/or analyzed in this study are available from the Lingbing Zeng (E-mail: [email protected]) and Junming Li (E-mail: [email protected]) on reasonable request.
Ethics Approval
This study was approved by the Ethics Committee of the first affiliated hospital of Nanchang University (Code: 2014056). All participants in this study provided written informed consent. This study was conducted in accordance with the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China (32060040 and 31760261), the National Science Foundation of Jiangxi province (20202BABL216084 and 20192ACBL21042), and the Science and Technology Plan Projects of Health and Family Planning Commission of Jiangxi province (20203692).
Disclosure
The authors declare no conflicts of interest.
References
1. Ko WC, Paterson DL, Sagnimeni AJ, et al. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis. 2002;8:160–166. doi:10.3201/eid0802.010025
2. Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12:881–887. doi:10.1016/S1473-3099(12)70205-0
3. Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–796. doi:10.1016/S1473-3099(13)70190-7
4. Slover CM, Rodvold KA, Danziger LH. Tigecycline: a novel broad-spectrum antimicrobial. Ann Pharmacother. 2007;41:965–972. doi:10.1345/aph.1H543
5. Zhong X, Xu H, Chen D, Zhou H, Hu X, Cheng G. First emergence of acrAB and oqxAB mediated tigecycline resistance in clinical isolates of Klebsiella pneumoniae pre-dating the use of tigecycline in a Chinese hospital. PLoS One. 2014;9:e115185. doi:10.1371/journal.pone.0115185
6. Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. doi:10.3389/fmicb.2016.00895
7. Cheng YH, Huang TW, Juan CH, et al. Tigecycline-non-susceptible hypervirulent Klebsiella pneumoniae strains in Taiwan. J Antimicrob Chemother. 2020;75:309–317. doi:10.1093/jac/dkz450
8. Xu H, Zhou Y, Zhai X, et al. Emergence and characterization of tigecycline resistance in multidrug-resistant Klebsiella pneumoniae isolates from blood samples of patients in intensive care units in northern China. J Med Microbiol. 2016;65:751–759. doi:10.1099/jmm.0.000299
9. Jeong SH, Kim HS, Kim JS, et al. Prevalence and molecular characteristics of carbapenemase-producing Enterobacteriaceae from five hospitals in Korea. Ann Lab Med. 2016;36:529–535. doi:10.3343/alm.2016.36.6.529
10. Hladicz A, Kittinger C, Zarfel G. Tigecycline resistant Klebsiella pneumoniae isolated from Austrian River Water. Int J Environ Res Public Health. 2017;14:1169. doi:10.3390/ijerph14101169
11. Chiu SK, Huang LY, Chen H, et al. Roles of ramR and tet(A) mutations in conferring tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2017;61. doi:10.1128/AAC.00391-17
12. Villa L, Feudi C, Fortini D, Garcia-Fernandez A, Carattoli A. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob Agents Chemother. 2014;58:1707–1712. doi:10.1128/AAC.01803-13
13. Moore IF, Hughes DW, Wright GD. Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry. 2005;44:11829–11835. doi:10.1021/bi0506066
14. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi:10.1128/JCM.43.8.4178-4182.2005
15. Gona F, Comandatore F, Battaglia S, et al. Comparison of core-genome MLST, coreSNP and PFGE methods for Klebsiella pneumoniae cluster analysis. Microb Genom. 2020;6. doi:10.1099/mgen.0.000347
16. Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–236. doi:10.1016/S1473-3099(09)70054-4
17. Neuner EA, Yeh JY, Hall GS, et al. Treatment and outcomes in carbapenem-resistant Klebsiella pneumoniae bloodstream infections. Diagn Microbiol Infect Dis. 2011;69:357–362. doi:10.1016/j.diagmicrobio.2010.10.013
18. Pournaras S, Koumaki V, Spanakis N, Gennimata V, Tsakris A. Current perspectives on tigecycline resistance in Enterobacteriaceae: susceptibility testing issues and mechanisms of resistance. Int J Antimicrob Agents. 2016;48:11–18. doi:10.1016/j.ijantimicag.2016.04.017
19. Wang X, Chen H, Zhang Y, et al. Genetic characterisation of clinical Klebsiella pneumoniae isolates with reduced susceptibility to tigecycline: role of the global regulator RamA and its local repressor RamR. Int J Antimicrob Agents. 2015;45:635–640. doi:10.1016/j.ijantimicag.2014.12.022
20. Veleba M, Higgins PG, Gonzalez G, Seifert H, Schneiders T. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2012;56:4450–4458. doi:10.1128/AAC.00456-12
21. Yu X, Zhang W, Zhao Z, et al. Molecular characterization of carbapenem-resistant Klebsiella pneumoniae isolates with focus on antimicrobial resistance. BMC Genomics. 2019;20:822. doi:10.1186/s12864-019-6225-9
22. Zeng L, Deng Q, Zeng T, Liu Y, Zhang J, Cao X. Prevalence of carbapenem-resistant Klebsiella pneumoniae infection in Southern China: clinical characteristics, antimicrobial resistance, virulence, and geographic distribution. Microb Drug Resist. 2020;26:483–491. doi:10.1089/mdr.2018.0401
23. Chung HS, Yong D, Lee M. Mechanisms of ertapenem resistance in Enterobacteriaceae isolates in a tertiary university hospital. J Investig Med. 2016;64:1042–1049. doi:10.1136/jim-2016-000117
24. Wu JJ, Wang LR, Liu YF, Chen HM, Yan JJ. Prevalence and characteristics of ertapenem-resistant Klebsiella pneumoniae isolates in a Taiwanese university hospital. Microb Drug Resist. 2011;17:259–266. doi:10.1089/mdr.2010.0115
25. Candan ED, Aksoz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62:867–874. doi:10.18388/abp.2015_1148
26. Yu WL, Ko WC, Cheng KC, et al. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42:1351–1358. doi:10.1086/503420
27. Liu Y, Liu PP, Wang LH, Wei DD, Wan LG, Zhang W. Capsular polysaccharide types and virulence-related traits of epidemic KPC-producing Klebsiella pneumoniae isolates in a Chinese University Hospital. Microb Drug Resist. 2017;23:901–907. doi:10.1089/mdr.2016.0222
28. Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66:307–312. doi:10.1093/jac/dkq431
29. Liu L, Feng Y, Tang G, et al. Carbapenem-resistant isolates of the Klebsiella pneumoniae complex in Western China: the common ST11 and the surprising hospital-specific types. Clin Infect Dis. 2018;67:S263–S265. doi:10.1093/cid/ciy662
30. Yan WJ, Jing N, Wang SM, et al. Molecular characterization of carbapenem-resistant Enterobacteriaceae and emergence of tigecycline non-susceptible strains in the Henan province in China: a multicenter study. J Med Microbiol. 2021;70. doi:10.1099/jmm.0.001325
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

