Back to Journals » International Journal of Nanomedicine » Volume 14
Modification of fliposomes with a polycation can enhance the control of pH-induced release
Authors Lokova AY, Zaborova OV
Received 9 October 2018
Accepted for publication 27 December 2018
Published 8 February 2019 Volume 2019:14 Pages 1039—1049
DOI https://doi.org/10.2147/IJN.S190306
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Anastasia Yu Lokova, Olga V Zaborova
Department of Chemistry, M.V. Lomonosov Moscow State University, 119991 Moscow, Russian Federation
Purpose: Nowadays, the development of stimuli-sensitive nanocontainers for targeted drug delivery is of great value. Encapsulation of a drug in a pH-sensitive liposomal container not only provides protective and transport functions, but also helps to create a system with a controlled release mechanism.
Methods: In this study, we investigated the influence of a cationic polypeptide on the pH-induced release of anticancer drug doxorubicin (DXR) from the anionic fliposomes – liposomes consisting of a neutral lipid, an anionic lipid (prone to interact with a polycation), and a lipid trigger (imparting the pH-sensitivity).
Results: First, we showed the possibility to control the pH-induced release by the simple modification of the anionic fliposomes with linear polylysine. Second, we optimized the fliposomal composition such that the obtained fliposomes responded to the pH changes only when complexed with the polycation (“turning on” the release). Finally, pH-induced release from the polylysine-modified anionic fliposomes was tested on an anticancer drug DXR.
Conclusion: We have succeeded in developing “smart” stimuli-sensitive nanocontainers capable of tunable controlled release of a drug. Moreover, based on the data on release of a low molecular salt, one can predict the release profile of DXR.
Keywords: polylysine, polymer–colloid complex, stimuli-sensitive nanocontainer, pH-sensitivity, controllable drug release
Introduction
The development of nanocontainers for the delivery of biologically active compounds has attracted considerable attention for its potential application in pharmacology. The encapsulation of biologically active compounds can bring a variety of benefits including a reduction in the side effect and an increase in the bioavailability of substances.1,2 Polymeric micelles,3,4 nanoemulsions,5 and liposomes6,7 are amongst the most studied systems to develop such containers. However, despite a significant progress in the design of liposomal nanocontainers, only a few of them are available on the market,8,9 mainly due to the low therapeutic effect of the liposomal formulations which is directly related to the low efficiency of drug release. It has been recognized now that the targeted release of the encapsulated compound is one of the approaches to increase the therapeutic efficiency, as a result of the environmental changes in the infected tissue such as pH,10 redox potential,11 enzyme activity,12 etc. For example, the tumor extracellular pH is slightly acidic in comparison to the blood pH,13 thus the containers capable of releasing the drug in response to a decrease of pH are of great interest.14–17
The vast majority of the pH-sensitive liposomal agents are weak acids or acid-hydrolyzed lipid derivatives,18 including the most widely used pH-sensitive liposomes with phosphatidylethanolamine (PE) and the complementary lipid, accessible for protonation.19–21 The complementary lipid stabilizes the mixed bilayer membrane at neutral pH, whereas it becomes protonated and immiscible with the PE molecules after decrease in pH resulting in disruption of the liposomes and formation of individual vesicles and hexagonal phase of PE lipids.22 Another method to prepare the pH-sensitive liposomes is to use molecules possessing weak acidic properties, for example, cholesteryl hemisuccinate (CHEMS), which has a negative charge at neutral pH and thus ensures the aggregative stability of the liposomes. Decrease of pH leads to the neutralization of the CHEMS molecule and causes the liposomal aggregations and the release of the encapsulated compound.23 Polyelectrolytes, such as polyacrylic acid24,25 or carboxylated hyaluronic acid,26 have also been used as pH-sensitive agents for liposomes, as they become hydrophilic with the decrease of pH value and, thus, disrupt a liposomal membrane. The formation of sterically stabilized polyethyleneglycol (PEG)–phenyl vinyl ether (PIVE)–lipid conjugate liposomes was recently reported.27 The acid-labile bond between PEG and PIVE lipid derivative cleaves under the acidic conditions, resulting in the loss of stability effect, liposome aggregation, and the release of content.
Conformational switches are another class of pH-sensitive liposomal agents that are represented by quasi-lipids based on cyclohexane,28 piperidine,29 or bispidinone.30 Trans-4,5-didodecyloxycarbonyl-2-morpholinocyclohexanol (TR)-based lipid31 is reported to belong to this class of agents and is the most promising for biomedical applications. Protonation leads to the formation of a hydrogen bond between morpholine and hydroxyl groups resulting in the conformational flip of a cyclohexanol ring and, finally, to the disruption of the liposomal membrane and the release of the encapsulated compound. The liposomes containing this type of a lipid molecule are called “fliposomes”.31 The pH-induced conformational flip of TR is presented in Figure 1.
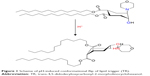  | Figure 1 Scheme of pH-induced conformational flip of lipid trigger (TR). |
This study aimed to develop a tunable pH-sensitive liposomal container for the encapsulation and controlled release of the anticancer drug doxorubicin (DXR). We investigated the possibility of controlling the pH-induced release of the encapsulated compounds by simple modification of fliposomes by using a linear biocompatible polycation. Previously, we studied the interaction of branched polycations (brush-like32 or star-shaped33) with anionic fliposomes and showed an increase in the rate and amount of the content release. However, the adsorption of the branched polycations induces the transmembrane migration (flip-flop) of the anionic lipids from the inner to the outer layer of a liposomal membrane,34 thus causing the formation of undesirable defects inside the membrane and limiting the application of such constructions for drug delivery. We decided to use linear polylysine, to prevent the formation of the undesirable defects upon adsorption of the polycation on the surface of anionic fliposomes as this polymer does not induce flip-flop of the anionic lipids.35 Moreover, polylysine is capable of undergoing enzymatic degradation due to its polypeptide structure and can also be used as a model to mimic blood proteins.
Materials and methods
Materials
α-Polylysine hydrobromide (pLys) with a chain length of 70 units (MW =15,000 g/mol) was purchased from Sigma Aldrich Co. (St Louis, MA, USA). Polylysine concentration is given as number of moles of the repeating units per liter. 1,2-Dioleoyl-sn-glycero-3-phosphocholine (PC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (PS), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (PE-Rhod) were purchased from Avanti Polar Lipids Inc (Alabaster, AL, USA). The chemical structures of the lipids used in the study are presented in Figure 2. TR (lipid trigger) was synthesized in N.D. Zelinsky Institute of Organic Chemistry by Vasily Migulin as described elsewhere.36
The buffer solutions used were as follows: Tris buffer ((HOCH2)3CNH2), 1 mmol/L, pH 7.4; phosphate buffer (Na2HPO4, NaH2PO4, NaCl), 10 mmol/L, pH 7.4; acetate buffer (CH3COOH, CH3COONa), 1 mmol/L, pH 5.5.
Methods
Preparation of liposomes
Small monolamellar liposomes were prepared using the standard procedure.37 Breifly, the required amounts of lipids were dissolved in chloroform. Then the organic solvent was removed under vacuum (Rotavapor; Buchi, Flawil, Switzerland) at 36°C. A thin lipid film was dispersed in a buffer solution. Then the dispersion was sonicated using a titanium probe for 2×300 seconds (Ultrasonic homogenizer 4710; Cole-Parmer, Niles, IL, USA). Obtained liposomes were centrifuged (Centrifuge J-11; Beckman, Palo Alto, CA, USA) for 5 minutes at 11,000 rpm to separate from titanium dust. The molar ratio of the lipids PS/TR/PC in the liposomes formed was 1/3/6 (fliposomes 1), 3/0.5/6.5 (fliposomes 2), and 3/1/6 (fliposomes 3). The lipid content, effective diameter (Deff), and the electrophoretic mobility (EPM) of the fliposomal formulations used in 1 mmol/L Tris buffer solution, pH 7.4, are presented in Table 1. The obtained liposomal suspension was stored at 4°C and used within 3 days.
Fluorescent-labeled liposomes were prepared using the same procedure, except that 0.5 wt% of PE-Rhod lipid (Figure 2) was added to the lipid mixture solution before chloroform evaporation.
NaCl-loaded liposomes were prepared by dispersing the lipid film in 1 M NaCl solution followed by the dialysis of the liposomal suspension against water as described previously.38 The obtained NaCl-loaded liposomes were stored at 4°C and used within 2 days.
Ammonium sulfate gradient liposomes for the DXR loading were prepared by dispersing the lipid film in 100 mmol/L (NH4)2SO4 solution and then dialyzing against PBS (10 mmol/L, pH=7.4).39 The DXR solution was added to the liposomal suspension in the ratio lipid/DXR =10/1 (w/w). The unloaded DXR was separated via Sepharose G-25 column using PBS as eluent. Liposomal suspension was stored at 4°C and used within 1 day.
Dynamic light scattering (DLS)
Effective diameters of liposomes and their complexes with polylysine were determined by DLS in a thermostated cell with a Brookhaven Zeta Plus instrument (New York, NY, USA). The intensity of the scattered light was detected at the angle θ=90° using a laser with a wavelength of 640 nm. DLS data were evaluated using the software provided by the manufacturer. The values were the average of at least three independent measurements.
Microelectrophoresis
The EPM was measured by laser microelectrophoresis technique40 using the same Brookhaven Zeta Plus instrument used for DLS measurements. The software provided by the manufacturer was used to calculate the EPM of the samples. Each mobility value presented in the text is an average of ten values.
Fluorescence
The fluorescence intensity of rhodamine-labeled liposome suspensions was measured at λem = 571 nm (λex = 557 nm) using F-4000 Hitachi fluorescence spectrofluorimeter (Hitachi, Japan). The release of DXR was monitored using F-4000 Hitachi fluorescence spectrofluorimeter (λem =555 nm, λex = 490 nm). The cell temperature was maintained constant at 25°C±0.1°C or 37°C±0.1°C.
Conductometry
The NaCl release was monitored by measuring the conductivity of the NaCl-loaded vesicle suspensions in a buffer solution with a CDM83 conductometer (Radiometer, Copenhagen, Denmark). The cell temperature was maintained constant at 25°C±0.1°C or 37°C±0.1°C.
Potentiometry
The pH values of buffer solutions were determined using the pH-meter 210 (Hanna, Woonsocket, RI, USA) with a glass electrode HI 1131B.
Results and discussion
First, the dependence of the degree of protonation of polylysine amino groups (α) as a function of pH was investigated using the method described previously.35 Polylysine is a weak base with a theoretical pKa,0 value about 10.44,41 thus a 0.4 mM water solution of pLys was converted into a free-base form by addition of an equimolar amount of NaOH and titrated with 0.1 M HCl solution. The equivalence point obtained from the titration curve (Figure 3A) was at pH=6.77. The pH dependence of α values was calculated as the second step based on the potentiometric titration data (Figure 3B). The interaction of polylysine with anionic fliposomes was studied further at pH 7.4 which corresponds to almost fully protonated form of the polycation.
Complexation of fliposomes 1 with linear polylysine
Addition of polylysine to the suspension of anionic fliposomes 1 led to the decrease of surface charge of the particles due to the formation of the polycation–liposomal electrostatic complex (Figure 4, curve 1). The complex formation was followed by the laser microelectrophoresis technique.
As one can see from Figure 4, the concentration of polylysine needed for the neutralization of the total surface charge of forming complexes was 0.056 mmol/L which corresponds to the anionic lipid/cationic groups ratio, [−]/[+]=2, suggesting that only a half of the anionic lipids present in the system interacted with polylysine. It was argued earlier that lipids are distributed uniformly between the inner and outer leaflets in the model membranes.42 Thus, the formation of the neutral complex at [−]/[+]=2 implies that polylysine interacts with the anionic lipids located only on the outer leaflet of the liposomal membrane. In other words, the adsorption of polylysine does not induce a flip-flop of anionic lipids, which is consistent with the previously published data.35 Further addition of polylysine resulted in the sign change of the EPM of particles. The size measurements of the complexes showed a common behavior for the polyion-decorated vesicles connected with the re-entrant condensation:43 the complex size increased with the approach of the particle charge neutralization point and size decrease after passing the charge inversion point (Figure 4, curve 2). The complexes with the excess of polylysine ([−]/[+]<1) were colloidally stable with surface charge value of 1.5 (μm/s)/(V/cm) and had the size of the initial liposomes.
Once the complex is formed, its association–dissociation stability should depend on the ionic strength of the media. The complex stability was evaluated by the titration of fluorescently-labeled fliposomes with polylysine solution.35,44 Formation of the complex led to the decrease of fluorescence intensity due to the static quenching45 (Figure 5A). Further addition of the NaCl solution to the formed complex resulted in an increase of the media ionic strength and, at a certain concentration, led to fluorescence recovery (Figure 5B), indicating the complex dissociation.
The formation of complexes was investigated at two different pH values corresponding to the pH of healthy tissues (pH=7.4, Tris buffer) and tumors (pH=5.5, acetate buffer). Figure 5A shows that the change in pH did not change the nature of the interaction between polylysine and anionic fliposomes, although the lipid trigger was partially protonated at pH=5.5.28 Nevertheless, we observed some differences in the salt concentration required for complex dissociation (Figure 5B). At pH 7.4, the complex was stable up to 0.2 M NaCl (Figure 5B, curve 1), while the complex started to dissociate at NaCl concentration higher than 0.05 M at pH 5.5 (Figure 5B, curve 2). The complex formed initially at pH 7.4 and when transferred to pH 5.5 (Figure 5B, curve 3) had the same stability as the complex formed at pH 5.5. Such differences in the NaCl concentration needed for the complex dissociation at pH=7.4 and pH=5.5 could be attributed to 1) different [−]/[+] ratios of the complexes titrated by NaCl solution – 1 and 0.77 for complexes formed at pH=7.4 and 5.5, respectively; 2) partial protonation of lipid trigger that facilitates the complex dissociation; and 3) probable influence of the nature of buffer ions on the dissociation of complexes. Nevertheless, the influence of the ionic strength on the dissociation stability of anionic fliposomes/polylysine complexes is out of scope of the current research.
Tunable pH-induced release – acceleration of the release rate (fliposomes 1)
The pH-induced release of the encapsulated hydrophilic compound was investigated using the NaCl-loaded fliposomes. The complexes with excess of polylysine ([−]/[+]=0.5) were used. The salt release was followed conductometrically46 at two different temperatures: 25°C (Figure 6A) and 37°C (Figure 6B). The control experiments at both temperatures (Figure 6, curve 3) showed that the adsorption of polylysine at pH 7.4 does not disturb the lipid membrane and thus does not lead to the release. The suspension of individual fliposomes or the complex formed at pH 7.4 was transferred to the buffer solution with pH 5.5 and the conductivity was monitored at different time intervals. The measured values were normalized to the conductivity of the system treated with Triton X-100; this state was attributed to the complete rupture of the fliposomes.
The change in pH induced the immediate onset of the release of contents from both the individual fliposomes (Figure 6A, curve 1) and fliposomes in the complex (Figure 6A, curve 2). The initial release rate from individual fliposomes (0.012 min−1) was ∼2.4 times lower than from the fliposomes in complex (0.029 min−1). The maximal release for fliposomes in complex (0.19) was 2.4 times higher than for individual fliposomes (0.08) and was observed 40 and 15 minutes after the change of pH for fliposomes in complex and individual fliposomes, respectively. The higher release rate from the fliposomes in the complex could be connected with an increase in the local concentration of the lipid trigger and a subsequent increase in the concentration of defects, due to the lateral segregation of the anionic lipids under the influence of the polycation. The leakage was not complete in both cases, similar to the systems described earlier using the same fliposomes with polycationic brushes32 and star-shaped polycations.33 We assume that “healing” of the defects in the membrane is responsible for the incomplete salt leakage. Indeed, in all cases the leakage reached a plateau about 0.5 hours after the change of pH both for individual fliposomes and fliposomes in complex with branched and linear polycations.
Interestingly, the maximal release from fliposomes in complexes with branched polycations32,33 was 60%, whereas in the system with linear polycation it was 20%. This finding corroborates with the occurrence of flip-flop of anionic lipids induced by the polycation adsorption. The flip-flop mediates an increase in the anionic lipid molar fraction in the outer layer of the liposomal membrane; the adsorption of polycation induces the lateral segregation of anionic lipids,34 and consequently, increase of the concentration of lipid trigger in the unaffected surface area. Thus the pH-induced release from the complexes of anionic fliposomes with polycation should be more effective in the system with flip-flop of anionic lipids, as the higher concentration of lipid trigger leads to the higher concentration of the defects formed in response to the change in pH.
The release at T=37°C (Figure 6B) had the same features as the release at T=25°C; the release rate from the individual fliposomes (0.062 min−1) was slower in comparison with fliposomes 1/polylysine complex (0.126 min−1). The increase of temperature resulted in five times increase of the initial release rate and, consequentially, in about two times increase in the maximal release – 0.26 for individual fliposomes 1 and 0.43 for fliposomes 1 in complex with polylysine.
We showed that we can influence the release rate of the encapsulated compound by modifying the anionic fliposomes with linear polylysine and forming a stable and biocompatible polymer–colloid complex after the decrease of pH. Thereby, it should also be possible to create the container that releases the inner content in response to the decrease of pH only if modified with the polycation.
Composition optimization (fliposomes 2 and 3)
We prepared the fliposomes with altered composition to investigate whether it is possible to make fliposomal container responsive to the pH decrease only in a complex with polycations. The content of anionic lipid was increased to a molar fraction of 30% to enhance the effect of lateral concentration of anionic lipids induced by the polycation. It was previously shown47 that the increase of the amount of anionic lipid up to the molar fraction of 30% improves the stability of the polycation–liposome complex in water-salt media, although the liposomal structure is not still disrupted under the adsorption of the polycation. Two molar fractions of lipid trigger were used: 5% for fliposomes 2 and 10% for fliposomes 3 to suppress the release from the individual fliposomes after decreasing of pH (Table 1).
The changes in the surface charge and size of particles are presented in Figure 7 as a function of the polylysine concentration. The neutralization of the fliposomal surface charge occurred at the concentration of 0.11 mmol/L of added polylysine both for the systems with fliposomes 2 (Figure 7A, curve 1) and fliposomes 3 (Figure 7B, curve 3). This amount corresponded to the [−]/[+]=2 and was attributed to the absence of flip-flop of the anionic lipid similar to fliposomes 1.
The stability of the fliposome/polylysine complex in water-salt media was evaluated as described for the system with fliposomes 1 (Figure 5). The addition of polylysine solution to the suspension of anionic fliposomes 3 led to complex formation irrespective of the pH of media (Figure 8A). The complexes were stable up to a salt concentration of 0.2 M (Figure 8B). The data for the fliposomes 2 system are not presented as they are identical to the data for the fliposomes 3 system (Figure 8). We conclude that the complexes of fliposomes 3 with polylysine seemed to be more stable in water-salt media than complexes of fliposomes 1. An increase in the stability of the anionic liposome–polycation complexes with the increase of the molar fraction of anionic lipid was also observed44,47,48 for the systems with branched polycations.
Tunable pH-induced release – “turning on” the release (fliposomes 2 and 3)
The pH-induced release was tested on the fliposomes 2 and fliposomes 3 loaded with 1 M NaCl both for individual fliposomes and fliposomes in complex with the excess of polycation at two temperatures: 25°C and 37°C. The leakage from individual fliposomes 2 was about 0.03 after 2 hours of pH change at both temperatures (Figure 9, curve 1). The fliposomes 2 in complex with polylysine released the encapsulated salt up to 0.04 and 0.06 at 25°C and 37°C, correspondingly (Figure 9, curve 2). The individual fliposomes 3 did not leak after pH change at 25°C (Figure 9A, curve 3), while there was a negligible release up to 0.03 at 37°C (Figure 9B, curve 3). We regard the salt leakage of 0.05 as negligible as it was comparable with the instrumental error. On the other hand, fliposomes 3 complex showed fourfold release of the inner content with the increase of temperature: 0.007 min−1 at 25°C and 0.029 min−1 at 37°C. The maximal release was 0.08 and 0.21 for the complexes at 25°C and 37°C, correspondingly (Figure 9, curve 4).
The release experiments (Figure 9) confirmed the possibility to govern the pH-induced release from the fliposomes by their modification with the linear polycation and the release dependence on the lipid trigger molar fraction. In the case of fliposomes 2, the amount of lipid trigger (5% molar) was insufficient for the effective disturbance of the lipid membrane after pH change. Thus, we chose the system with fliposomes 3 for the next experiment. Herein, we want to address the following question: Can the above-mentioned approach be applicable for the fliposome system loaded with a real drug? The pH-induced release of anticancer drug DXR from fliposomes 3 was further investigated for that purpose.
Tunable pH-induced release – doxorubicin release (fliposomes 3)
DXR was incorporated into the fliposomes by remote loading driven by ammonium sulfate gradient (see the details in Methods section). Acidic pH is not required for liposomal preparation by this method,49–51 which is critical for the formation of liposomes containing the pH-sensitive agent in the membrane. The difference in ammonium concentration in the inner and outer volume of liposomes induces a transmembrane pH-gradient with the intraliposomal pH=5.5.49 We have shown (Figures 6 and 9) that the decrease in pH value to 5.5 can induce defect formation in the fliposomal membrane and this can cause a decrease in ammonium gradient during the formation of fliposomes. Thus, we have used fliposomes 3, as the individual fliposomes in this case showed no release at pH=5.5 (Figure 9A, curve 1), while the modification of those fliposomes with polylysine led to the release of the encapsulated salt (Figure 9, curve 2). DXR accumulated in the liposomal inner volume due to the formation of a gel-like precipitate of DXR-sulfate salt inside the liposomes. The precipitation of DXR does not reduce its bioavailability.49
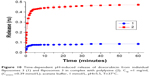
The pH-induced release was followed by the increase of fluorescence intensity due to the increase of DXR concentration in the inner solution (Figure 10). This experiment was conducted in PBS with 0.15 M NaCl at 37°C. The decrease of pH value to 5.5 induced the immediate release of anticancer drug both from individual fliposomes and fliposomes in complex. Most part of the drug was released within the first 5 minutes after pH change. The maximal release was 0.08 and 0.47 of the loaded drug for individual fliposomes 3 and fliposomes 3 in complex, correspondingly. These data correlated very well with the release of salt from the same fliposomes (Figure 9B), although the mechanism of the release of salt and DXR is different. The pH-induced lipid trigger flip is responsible for the salt release through the pores in the liposomal membrane. DXR release is more complicated since it is affected by several factors: phase state of the lipid bilayer, permeability of the liposomal membrane, and the solubility of the drug encapsulated into the liposomes.52 The major mechanism of DXR release is diffusion of the deprotonated drug through the liposomal membrane via dissolution–diffusion mechanism; thus, an increase in membrane permeability results in an increase in release rate.53 The decrease of outer pH value to 5.5 induces the flip of lipid trigger and formation of the defects that increases the permeability of the membrane and facilitates the leakage of DXR from the fliposomes. As the defects are also permeable for the buffer components, it leads to the disruption of the buffer transmembrane gradient and deprotonation of DXR inside the liposomes.
Conclusion
We have investigated the influence of the linear biocompatible polylysine on the pH-induced release from the anionic fliposomes at two different temperatures (25°C and 37°C). As the adsorption of polylysine on the anionic fliposomes did not induce flip-flop of anionic lipids, the liposomal membrane was not disturbed and polylysine did not cause the release at neutral pH. The pH responsivity of the fliposomes was achieved by embedding of the lipid trigger into the liposomal membrane. The amount of lipid trigger affects the ability of fliposomes to release the contents after the pH change. The salt release from the fliposomes with lipid trigger molar fraction 0.3 was up to 0.26 at 37°C, while from the fliposomes with TR molar fraction 0.05 and 0.1 showed only a slight salt release (up to 0.03) at the same temperature. We demonstrated that the modification of fliposomes with linear polylysine can both accelerate (fliposomes 1) and “turn on” (fliposomes 3) the release of encapsulated compound after decreasing the pH. The model salt release experiments can predict the release of the encapsulated drug. Moreover, the obtained data can help to predict the specificity of the pH-induced release from the fliposomes in physiological media in the presence of blood proteins.
Acknowledgments
This research was partially supported by the Russian Science Foundation (project 18-73-00076). Dr Vasily A Migulin is acknowledged for the synthesis of the lipid trigger. The authors express gratitude to the team of the Laboratory of Nanobiostructures of the Polymer Chair of the Chemistry Department of Lomonosov Moscow State University for fruitful discussions and constructive comments and especially to Prof Alexander A Yaroslavov for invaluable support.
Disclosure
The authors report no conflicts of interest in this work.
References
Wang S, Su R, Nie S, et al. Application of nanotechnology in improving bioavailability and bioactivity of diet-derived phytochemicals. J Nutr Biochem. 2014;25(4):363–376. | ||
Watkins R, Wu L, Zhang C, Davis R, Xu B. Natural product-based nanomedicine: recent advances and issues. Int J Nanomed. 2015;10(1):6055–6074. | ||
Miyata K, Christie RJ, Kataoka K. Polymeric micelles for nano-scale drug delivery. React Funct Polym. 2011;71(3):227–234. | ||
Gong J, Chen M, Zheng Y, Wang S, Wang Y. Polymeric micelles drug delivery system in oncology. J Control Release. 2012;159(3):312–323. | ||
Singh Y, Meher JG, Raval K, et al. Nanoemulsion: concepts, development and applications in drug delivery. J Control Release. 2017;252:28–49. | ||
Allen TM, Cullis PR. Liposomal drug delivery systems: from concept to clinical applications. Adv Drug Deliv Rev. 2013;65(1):36–48. | ||
Sercombe L, Veerati T, Moheimani F, Wu SY, Sood AK, Hua S. Advances and challenges of liposome assisted drug delivery. Front Pharmacol. 2015;6(127):286. | ||
Torchilin VP. Recent advances with liposomes as pharmaceutical carriers. Nat Rev Drug Discov. 2005;4(2):145–160. | ||
Cheung C, Al-Jamal WT. Liposomes-based nanoparticles for cancer therapy and bioimaging. In: Gonçalves G, Tobias G, editors. Nanooncology Nanomedicine and Nanotoxicology. Cham: Springer; 2018:51–87. | ||
Mo R, Sun Q, Xue J, et al. Multistage pH-responsive liposomes for mitochondrial-targeted anticancer drug delivery. Adv Mater. 2012;24(27):3659–3665. | ||
Noyhouzer T, L’Homme C, Beaulieu I, et al. Ferrocene-modified phospholipid: an innovative precursor for redox-triggered drug delivery vesicles selective to cancer cells. Langmuir. 2016;32(17):4169–4178. | ||
Hu Q, Katti PS, Gu Z. Enzyme-responsive nanomaterials for controlled drug delivery. Nanoscale. 2014;6(21):12273–12286. | ||
Tian L, Bae YH. Cancer nanomedicines targeting tumor extracellular pH. Colloids and Surfaces B: Biointerfaces. 2012;99:116–126. | ||
Staff RH, Gallei M, Landfester K, Crespy D. Hydrophobic nanocontainers for stimulus-selective release in aqueous environments. Macromolecules. 2014;47(15):4876–4883. | ||
Dai L, Zhang Q, Li J, Shen X, Mu C, Cai K. Dendrimerlike mesoporous silica nanoparticles as pH-responsive nanocontainers for targeted drug delivery and bioimaging. ACS Appl Mater Interfaces. 2015;7(13):7357–7372. | ||
Zhao G, Long L, Zhang L, et al. Smart pH-sensitive nanoassemblies with cleavable PEGylation for tumor targeted drug delivery. Sci Rep. 2017;7(1):3383. | ||
Filippov SK, Vishnevetskaya NS, Niebuur B-J, et al. Influence of molar mass, dispersity, and type and location of hydrophobic side chain moieties on the critical micellar concentration and stability of amphiphilic HPMA-based polymer drug carriers. Colloid Polym Sci. 2017;295(8):1313–1325. | ||
Drummond DC, Zignani M, Leroux J. Current status of pH-sensitive liposomes in drug delivery. Prog Lipid Res. 2000;39(5):409–460. | ||
Collins D, Litzinger DC, Huang L. Structural and functional comparisons of pH-sensitive liposomes composed of phosphatidylethanolamine and three different diacylsuccinylglycerols. Biochimica et Biophysica Acta (BBA) – Biomembranes. 1990;1025(2):234–242. | ||
Sudimack JJ, Guo W, Tjarks W, Lee RJ. A novel pH-sensitive liposome formulation containing oleyl alcohol. Biochim Biophys Acta Biomembr. 2002;1564(1):31–37. | ||
Sánchez M, Aranda FJ, Teruel JA, Ortiz A. New pH-sensitive liposomes containing phosphatidylethanolamine and a bacterial dirhamnolipid. Chem Phys Lipids. 2011;164(1):16–23. | ||
Fan Y, Chen C, Huang Y, Zhang F, Lin G. Study of the pH-sensitive mechanism of tumor-targeting liposomes. Colloids Surf B Biointerfaces. 2017;151:19–25. | ||
Duan Y, Wei L, Petryk J, Ruddy TD. Formulation Characterization and tissue distribution of a novel pH-sensitive long-circulating liposome-based theranostic suitable for molecular imaging and drug delivery. Int J Nanomed. 2016;11:5697–5708. | ||
Jelezova I, Drakalska E, Momekova D, et al. Curcumin loaded pH-sensitive hybrid lipid/block copolymer nanosized drug delivery systems. Eur J Pharm Sci. 2015;78:67–78. | ||
Simões MG, Alves P, Carvalheiro M, Simões PN. Stability effect of cholesterol-poly(acrylic acid) in a stimuli-responsive polymer-liposome complex obtained from soybean lecithin for controlled drug delivery. Colloids Surf B Biointerfaces. 2017;152:103–113. | ||
Miyazaki M, Yuba E, Hayashi H, Harada A, Kono K. Hyaluronic acid-based pH-sensitive polymer-modified liposomes for cell-specific intracellular drug delivery systems. Bioconjug Chem. 2018;29(1):44–55. | ||
Kim HK, van den Bossche J, Hyun SH, Thompson DH. Acid-triggered release via dePEGylation of fusogenic liposomes mediated by heterobifunctional phenyl-substituted vinyl ethers with tunable pH-sensitivity. Bioconjug Chem. 2012;23(10):2071–2077. | ||
Zheng Y, Liu X, Samoshina NM, Samoshin VV, Franz AH, Guo X. Fliposomes: trans-2-aminocyclohexanol-based amphiphiles as pH-sensitive conformational switches of liposome membrane – a structure-activity relationship study. Chem Phys Lipids. 2018;210:129–141. | ||
Samoshin AV, Joo H, Korneichuk AY, Veselov IS, Grishina GV, Samoshin VV. Trans-3-Hydroxy-4-morpholinopiperidine – the pH-triggered conformational switch with a double FLIP. Tetrahedron Letters. 2013;54(8):1020–1024. | ||
Veremeeva PN, Bovina EM, Grishina IV, Lapteva VL, Palyulin VA, Zefirov NS. Synthesis of amphiphilic diacyl derivatives of 3,7-diazabicyclo[3.3.1]nonan-9-one. Mendeleev Communications. 2018;28(1):25–26. | ||
Samoshina NM, Liu X, Brazdova B, Franz AH, Samoshin VV, Guo X. Fliposomes: pH-sensitive liposomes containing a trans-2-morpholinocyclohexanol-based lipid that performs a conformational flip and triggers an instant cargo release in acidic medium. Pharmaceutics. 2011;3(3):379–405. | ||
Yaroslavov AA, Sybachin AV, Zaborova OV, et al. Capacious and programmable multi-liposomal carriers. Nanoscale. 2015;7(5):1635–1641. | ||
Sybachin AV, Zaborova OV, Imelbaeva KM, et al. Effects of the electrostatic complexation between anionic pH-sensitive liposomes and star-shaped polycations on the release of the liposomal content. Mendeleev Communications. 2016;26(4):276–278. | ||
Yaroslavov AA, Sybachin AV, Zaborova OV, et al. Multi-liposomal containers. Adv Colloid Interface Sci. 2015;226(Pt A):54–64. | ||
Yaroslavov AA, Kuchenkova OY, Okuneva IB, et al. Effect of polylysine on transformations and permeability of negative vesicular membranes. Biochimica et Biophysica Acta (BBA) – Biomembranes. 2003;1611(1–2):44–54. | ||
Brazdova B, Zhang N, Samoshin VV, Guo X. Trans-2-Aminocyclohexanol as a pH-sensitive conformational switch in lipid amphiphiles. Chem Commun. 2008;46(39):4774–4776. | ||
New RRC. Liposomes: A Practical Approach. Oxford: Oxford University Press; 1990. | ||
Zaborova OV, Filippov SK, Chytil P, et al. A novel approach to increase the stability of liposomal containers via in prep coating by poly[n-(2-hydroxypropyl)methacrylamide] with covalently attached cholesterol groups. Macromol Chem Phys. 2018:219(7):1700508. | ||
Alyane M, Barratt G, Lahouel M. Remote loading of doxorubicin into liposomes by transmembrane pH gradient to reduce toxicity toward H9c2 cells. Saudi Pharm J. 2016;24(2):165–175. | ||
Delgado AV, González-Caballero F, Hunter RJ, Koopal LK, Lyklema J. Measurement and interpretation of electrokinetic phenomena. Pure Appl Chem. 2005;77(10):1753–1805. | ||
Katchalsky A, Shavit N, Eisenberg H. Dissociation of weak polymeric acids and bases. J Polym Sci. 1954;13(68):69–84. | ||
Marquardt D, Geier B, Pabst G. Asymmetric lipid membranes: towards more realistic model systems. Membranes. 2015;5(2):180–196. | ||
Sennato S, Bordi F, Cametti C, Marianecci C, Carafa M, Cametti M. Hybrid niosome complexation in the presence of oppositely charged polyions. J Phys Chem B. 2008;112(12):3720–3727. | ||
Zaborova OV, Sybachin AV, Ballauff M, Yaroslavov AA. Structure and properties of complexes of polycationic brushes with anionic liposomes. Polym Sci Ser A. 2011;53(11):1019–1025. | ||
Lakowicz JR. Principles of Fluorescence Spectroscopy. Boston, MA: Springer; 2006. | ||
Bordi F, Cametti C, Sennato S, Viscomi D. Conductometric evidence for intact polyion-induced liposome clusters. J Colloid Interface Sci. 2006;304(2):512–517. | ||
Sybachin AV, Zaborova OV, Orlov VN, et al. Complexes between anionic liposomes and spherical polycationic brushes. an assembly of assemblies. Langmuir. 2014;30(9):2441–2447. | ||
Sybachin AV, Zaborova OV, Pergushov DV, et al. Complexes of star-shaped cationic polyelectrolytes with anionic liposomes: towards multi-liposomal assemblies with controllable stability. Polymer. 2016;93:198–203. | ||
Bolotin EM, Cohen R, Bar LK, et al. Ammonium sulfate gradients for efficient and stable remote loading of amphipathic weak bases into liposomes and ligandoliposomes. J Liposome Res. 1994;4(1):455–479. | ||
Fritze A, Hens F, Kimpfler A, Schubert R, Peschka-Süss R. Remote loading of doxorubicin into liposomes driven by a transmembrane phosphate gradient. Biochim Biophys Acta Biomembr. 2006;1758(10):1633–1640. | ||
Rehman AU, Omran Z, Anton H, et al. Development of doxorubicin hydrochloride loaded pH-sensitive liposomes: investigation on the impact of chemical nature of lipids and liposome composition on pH-sensitivity. Eur J Pharm Biopharm. 2018;133:331–338. | ||
Shibata H, Izutsu K, Yomota C, Okuda H, Goda Y. Investigation of factors affecting in vitro doxorubicin release from PEGylated liposomal doxorubicin for the development of in vitro release testing conditions. Drug Dev Ind Pharm. 2015;41(8):1376–1386. | ||
Farzaneh H, Ebrahimi Nik M, Mashreghi M, Saberi Z, Jaafari MR, Teymouri M. A study on the role of cholesterol and phosphatidylcholine in various features of liposomal doxorubicin: from liposomal preparation to therapy. Int J Pharm. 2018;551(1–2):300–308. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.