Back to Journals » OncoTargets and Therapy » Volume 12
miR520a-3p suppresses cell proliferation and metastasis by inhibiting the p65–NFκB pathway in glioblastoma
Received 15 March 2019
Accepted for publication 18 July 2019
Published 14 August 2019 Volume 2019:12 Pages 6503—6513
DOI https://doi.org/10.2147/OTT.S208889
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Jing-quan Zhang, Bo Hong
Department of Neurosurgery, Changhai Hospital, Second Military Medical University, Shanghai 200433, People’s Republic of China
Background: miR520a-3p has previously had its antitumorigenic role in various types of cancers revealed, and been predicted as a posttranscriptional regulator of the NFκB-subunit RELA gene. Thus, miR520a-3p could function in carcinogenesis through suppressing RELA.
Methods: Expression of miR520a-3p and RELA mRNA was quantified in glioma and normal tissue, and the correlation between them was analyzed statistically. Also, receiver-operating characteristic (ROC)–curve analysis was performed. Effects of miR520a-3p on cell viability, colony formation, migration, and invasion wereexplored in vitro. Whether RELA was a direct target of miR520a-3p or not was analyzed. Finally, restoration of RELA on the effect of miR520a-3p overexpression on proliferation of glioblastoma cells was detected.
Results: Data showed that miR520a-3p expression was aberrantly downregulated and associated with malignance in glioma tissue. Areas under ROC curves of miR520a-3p and RELA mRNA expression were 0.9483 and 0.5967, respectively. Also, miR520a-3p expression was statistically correlated with RELA mRNA level in grade III–IV glioma tissue. Transfection of miR520a-3p mimic significantly increased miR520a-3p expression, and resulted in significant suppression of proliferation, migration, and invasion of glioblastoma cells in vitro. miR520a-3p overexpression resulted in statistical downregulation of RELA, both in mRNA and protein levels. RELA was direct target of miR520a-3p. In addition, restoration of RELA significantly weakened the inhibitory effect of miR520a-3p overexpression on viability and EdU-labeled glioblastoma cells.
Conclusion: These findings suggest that miR520a-3p should be helpful in auxiliary glioma diagnosis and can attenuate the proliferation and metastasis of glioblastoma through suppressing RELA, and thus could be an attractive therapeutic target to eliminate glioblastoma.
Keywords: miR520a-3p, RELA, proliferation, migration, invasion, glioblastoma
Introduction
Gliomas are the most frequent malignant tumor the of central nervous system. According to microscopic similarities with putative cells of origin along glial precursor–cell lineages, gliomas are traditionally classified into two principal subgroups: diffuse gliomas characterized by extensive infiltrative growth into surrounding central nervous system parenchyma, and nondiffuse gliomas showing a more circumscribed growth pattern.1 In adults, the most prevalent glioma is glioblastoma, which can evolve rapidly over several weeks or months.2,3 Despite receiving surgery, radiotherapy, and chemotherapy, patients suffering from glioblastoma have an average survival time of only approximately 15 months.4 Therefore, complete understanding of molecular mechanisms underlying carcinogenesis of glioblastoma and other types of glioma is urgently needed, in order to provide a scientific basis for biomarkers and efficient therapies.
NFκB is an important transcription-factor family that contains five subunits: Rel (cREL), p65 (RELA, NFκB3), RELB, p105/p50 (NFκB1), and p100/p52 (NFκB2)5–7 All five subunits comprise a conserved REL homology domain (RHD) near the N-terminus. The RHD is crucial for dimerization of NFκB family members. The N-terminal part of the RHD is essential for DNA binding, while the C-terminal part is essential for interaction with inhibitors of κB. In addition, the subunits RELA, RELB, and cREL contain a C-terminal transactivation domain responding for transcriptional activation.8,9 NFκB family members can form homo- or heterodimers via the RHD. The p50 and p52 homodimers cannot activate gene transcription, but act as inhibitory molecules. RELB can form dimers only with p50 or p52, but other subunits can form either homodimers or heterodimers. Actually, the most common NFκB dimer is the p50–p65 heterodimer. Activation of NFκB signaling ultimately results in the translocation of released homodimers and/or heterodimers (canonical, p50–p65 heterodimer; uncanonical, p52–RELB heterodimer) into the nucleus and binds to κB elements located in distinct target genes to regulate gene transcription. Although the NFκB signal has recently had its tumor-suppressive role in certain cancers revealed, NFκB-pathway dysregulation occurs in both solid and hematopoietic malignancies and thus exerts a protumorigenic effect.10 As such, the NFκB pathway has been considered an attractive therapeutic target in various types of cancers, including glioblastoma.11,12 miR520a-3p has been found to be a tumor suppressor in various types of cancers. Also, miR520a-3p has been predicted as a posttranscriptional regulator of the NFκB-subunit RELA gene through online algorithm analysis, meaning that miR520a-3p can exert its role in carcinogenesis through suppressing RELA. Here, data showed that miR520a-3p was aberrantly downregulated in glioma tissue, and downregulation of miR520a-3p associated with malignance of glioma tissue and correlated with expression of RELA mRNA in World Health Organization (WHO) grade III–IV glioma tissue.Overexpression of miR520a-3p suppressed proliferation, migration, and invasion of glioblastoma cells in vitro and resulted in significant downregulation of RELA. RELA was a direct target of miR520a-3p. Restoration of RELA significantly weakened the inhibitory effect of miR520a-3p overexpression on proliferation of glioblastoma cells. These findings suggested that miR520a-3p should attenuate growth of glioblastoma cells through suppressing RELA/p65, and thus could be an attractive biomarker in auxiliary glioma diagnosis and a therapeutic target to eliminate glioblastoma.
Methods
Tissue collection
This study was approved by the ethics committee of Changhai Hospital, Second Military Medical University. A total of 30 glioma specimens and ten normal brain-tissue samples were obtained from Changhai Hospital of Second Military Medical University between May 2014 and June 2016. The study was conducted in accordance with the Declaration of Helsinki. All tumor tissue was excised from patients suffering from glioma and diagnosed using histopathology in accordance with WHO stage and grading system. Normal brain tissue was provided by patients who did not have cancer, but had suffered a traumatic brain injury and undergone subsequent surgical resection. Written informed consent was obtained from patients prior to the study. All tissue samples were immediately snap-frozen and stored in liquid nitrogen until use. Clinicopathological features of the 30 glioma patients are listed in Table S1.
Reverse-transcription quantitative PCR
Total RNA of tissue and cells was extracted using Trizol regent (15596018; Thermo Fisher Scientific) according to the manufacturer’s instructions, then purity detected using a NanoDrop2000 spectrophotometer (Thermo Fisher Scientific). RNA samples with high purity (OD260/OD2801.7–2.1) were used to reverse-transcribe cDNA using a PrimeScript RT reagent kit with gDNA Eraser (Perfect Real Time, RR047A; Takara) according to the manufacturer’s protocols. MicroRNAs were purified with a -SanPrep Column microRNA extraction kit (B518811; Sangon Biotech), and microRNA first-strand cDNA synthesis performed (tailing reaction, B532451; Sangon Biotech) according to the manufacturer’s instructions. Then, PCR amplification was carried out on a real-time PCR instrument (Slan 96P; Shanghai Hongshi Medical Technology) using AceQzR qPCR SYBRR green master mix (Q111-02; Vazyme Biotech) with cDNA as template. The reaction mixture comprised 10 μL 2× AceQ qPCR SYBR green master mix, 0.5 μL primer F (10 μM), 0.5 μL primer R (10 μM), 1 μL template DNA, and 8 μL double-distilled H2O. Raction conditions were predenaturation at 95°C for 5 minutes, followed by 40 cycles at 95°C for 10 seconds and 60°C for 30 seconds, and then 95°C for 15 seconds, 60°C for 60 seconds, and 95°C for 15 seconds. In this experiment, three equilibrium samples were set for each sample. Sequences of specific primers are listed in Table 1.
 |
Table 1 Reverse-transcription quantitative PCR primer sequences |
Cell culture
Human glioblastoma U251 and U87MG cell lines were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM (SH30022.01; HyClone) with 10% FBS (Thermo Fisher Scientific) and maintained at 37°C in a 5% CO2 humidified atmosphere.
Cell transfection
The hsa-miR520a-3p mimic (sense 5ʹ-AAAGUGCUUCCCUUUGGACUGU-3ʹ, antisense 5ʹ- AGUCCAAAGGGAAGCACUUUUU-3ʹ) and negative-control (NC) mimic (sense 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ, antisense 5ʹ-ACGUGACACGUUCGGAGAATT-3ʹ) were purchased from GenePharma (Shanghai, China). Transfection of RNA oligo was carried out using XtremeGene siRNA-transfection reagent (04476093001; Roche) according to the manufacturer’s instructions. Briefly, glioblastoma cells were harvested by trypsinization and then resuspended in six-well plates at a concentration of 5×105/well. Cells were incubated in DMEM with 10% FBS at 37°C until confluence reached 60%, then cultured in DMEM containing transfection working solution (500 μL/well) for 5 hours. Subsequently, the medium containing the transfection working solution was replaced with DMEM with 10% FBS without antibiotics, and cells were cultured in vitro for 48 hours before following experiments. Glioblastoma cells that stably overexpressed miR520a-3p were submitted to transfection with a recombinant vector containing human RELA using Lipofectamine 2000 (11668019; Thermo Fisher Scientific) according to the manufacturer’s instructions to restore RELA expression.
Plasmid construction and dual-luciferase reporter assays
To construct wild and mutated pmirGLO-RELA 3ʹUTR, PCR-amplification primers were designed by Primer 3 (version 0.4.0; http://frodo.wi.mit.edu) according to the sequence of human RELA3ʹUTR (gene ID 5970). Primer sequences are listed in Table 2. XhoI and SalI restriction-enzyme sites were introduced in the upstream of forward primers and downstream of reverse primers, respectively. With cDNA of 293T cells as template, PCR was performed (KOD FX 101; Toyobo Biotechnology) according to the manufacturer’s instructions. After being digested with XhoI (FD0694; Thermo Fisher Scientific) and SalI (FD0644; Thermo Fisher Scientific), wild and mutated human RELA 3ʹUTR gene fragments and pmirGLO dual firefly–Renillaluciferase miRNA target–expression vector (E1330; Promega Biotech) were ligated with Assembly Enzyme (TM201; ThinkGene Biotech) following the manufacturer’s protocols. Recombinant plasmids bearing wild and mutated human RELA 3ʹUTR were verified by sequencing.
 |
Table 2 PCR primers for amplification of wild and mutated human RELA 3ʹUTR |
Dual luciferase activity–reporter assays were performed using the R reporter assay system (E1910; Promega Biotech) according to the manufacturer’s instructions. In brief, 293T cells were cotransfected transiently with miRNA mimic and recombinant plasmids for 24 hours. Then, the cell-culture medium was removed and cells washed twice with PBS. Subsequently, cells were treated with passive lysis buffer for 20 minutes according to the protocol. The lysate was transferred to a 96-well plate, then the plate was placed in a SpectraMax M4 luciferase detector to detect firefly-luciferase and Renilla luciferase activity. The ratio of firefly-luciferase activity to Renilla luciferase activity was used to quantify luciferase activity.
Cell-viability assays
Cell viability was determined using CCK8 (C0038; Beyotime Biotechnology, Haimen, China) according to the manufacturer’s instructions. In brief, differently treated cells were seeded into 96-well plates at 105 cells/well. CCK8 was added to cell medium at various time points, then cells were incubated for 2 hours. OD450 was measured, and absorbance was an indicator of cell survival.
Colony-formation assays
Colony-formation assays were carried out to evaluate cell proliferation. Briefly, differently treated cells were harvested and seeded into six-well plates at a concentration of 1,500 cells/well and cultured in DMEM supplemented with 10% FBS. Two weeks later, visible colonies were fixed with 4% paraformaldehyde and underwent Giemsa staining for 20 minutes. Colonies (≥50 cells per colony) were counted and photographed.
5-Ethynyl-2ʹ-deoxyuridine–incorporation assays
EdU-incorporation assays were performed to visualize proliferating cells according to the manufacturer’s instructions (Cell-Light EdU Apollo 488 in vitro imaging kit, C10310-3; Guangzhou RiboBio). In brief, differently treated cells were seeded in triplicate in 96-well plates at a density of 5×103 cells/well and incubated in vitro for 48 hours. Then, 50 μmol/L EdU was added to the culture medium and cells incubated at 37°C for 4 hours. Subsequently, cells were treated with 4% paraformaldehyde for 30 minutes and 0.5% Triton X-100 for 20 minutes. After being rinsed with PBS three times, cells were exposed to 100 μL Appollo reaction cocktail for 30 minutes and incubated with 5 μg/mL Hoechst 33342 to label nuclei for 30 minutes. EdU-labeled cells and Hoechst 33342–stained cells were counted in ten random-view fields using fluorescent microscopy (Olympus IX71). Proliferating cells were calculated as the number of EdU-positive cells/the number of Hoechst-labeled cells.
Transwell invasion assays
Cell invasion was assessed using Corning BioCoat Matrigel invasion chambers according to the manufacturer’s instructions. Differently treated cells were harvested and resuspended in serum-free DMEM at adensity of 105 cells/well and placed into the 100 μL Matrigel–precoated upper chamber. The lower chamber was filled with DMEM supplemented with 20% FBS as a chemoattractant. After incubation for 48 hours, cells that had invaded through the 8 μm membrane were fixed with 4% paraformaldehyde for 15 minutes. Cells were then stained with 0.1% crystal violet and counted in five random fields using Image-Pro Plus.
Scratch assays
Cells undergoing different treatments were cultured in six-well plates until confluence. Then, cell layers were carefully wounded using sterile pipette tips and images of wound widths captured and quantified at 0 and 48 hours.
Western blotting
Cells were rinsed with ice-cold PBS and then lysed on ice with RIPA lysis buffer (PP110; Protein Biotechnology) according to the manufacturer’s instructions. Then, protein concentrations were determined using a BCA protein-assay kit (PP202; Protein Biotechnology) and denatured by boiling. Proteins were separated by 12% SDS-PAGE, then transferred onto a polyvinylidene fluoride membrane. The membrane was treated with Tris-buffered saline containing 0.1% Tween 20 and 5% nonfat milk for 1 hour. Then, the polyvinylidene fluoridemembrane was incubated with primary antibodies against p65 (ab32536; Abcam) and β-actin (ab115777; Abcam) overnight and then incubated with goat antirabbit IG (HRP) (ab205718; Abcam) for 2 hours at room temperature. Protein bands were visualized using a chemiluminescence kit (PP404; Protein Biotechnology).
Bioinformatic analysis
Potential miRNAs able to target human RELA were analyzed using TargetScan 5.1 (www.targetscan.org).
Statistical analysis
Data are presented as means ± SD and analyzed with SPSS 21.0 . One way ANOVAs or two-tailed Student’s t-tests were used to analyze differences between groups. The correlation of miR520a-3p expression with RELA mRNA was analyzed with Pearson’s correlation analysis. ROC-curve analysis was performed to evaluate the clinical value of miR520a-3p and RELA mRNA expression in glioma. P<0.05 was considered statistically significant.
Results
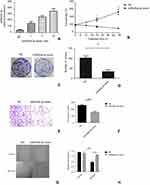
Downregulation of miR520a-3p was associated with malignancy and correlated with RELA mRNA in glioma
miR520a-3p and RELA mRNA expression in normal brain tissue and glioma tissue was analyzed using PCR. Compared to normal brain tissue, miR520a-3p was downregulated in graded glioma tissue diagnosed according to WHO classification. The average miR520a-3p level in grade III glioma tissue was lower than in grade II and higher than in grade IV (Figure 1A). miR520a-3p in glioma tissue from female patients was higher than from males (Figure 1B). ROC curves of miR520a-3p expression between normal brain tissue and gliomas (grade II–IV) showed that the area under the ROC curve was 0.9483 (95% CI 0.8852–1.011, P<0.0001; Figure 1C). On the other hand, endogenous RELA mRNA was detected in normal brain tissue. Expression of RELA mRNA in grade II tissue was slightly lower than in normal brain tissue, while RELA mRNA levels in grade III tissue was slightly higher than in normal tissue. Significant upregulation of RELA mRNA was observed in grade IV glioblastoma tissue compared to normal brain tissue (Figure 1D). Sex differences in RELA mRNA expression were not detected (Figure 1E). RELA mRNA expression between normal brain tissue and gliomas (grade II–IV) showed an area under the ROC curve of 0.5967 (95% CI 0.4185–0.7748, P>0.05; Figure 1F), indicating that RELA mRNA expression was relatively feeble in auxiliary glioma diagnosis compared with miR520a-3p expression. Nevertheless, expression of miR520a-3p was correlated with RELA mRNA expression in grade III–IV glioma tissue (Figure 1G)
Overexpression of miR520a-3p suppressed proliferation, migration, and invasion of glioblastoma cells in vitro
In order to understand the role of miR520a-3p on the carcinogenesis of glioblastoma, miR520a-3p in glioblastoma cells was reinforced by transfection with miR520a-3p mimics or the NC mimic. Then, differently treated cells were assessed for viability, clone formation, migration, and invasion. Data showed that transfection of miR520a-3p mimics generated upregulation of miR520a-3p in U251 cells in a dose-dependent manner (Figure 2A). Cells transfected with the NC mimic displayed a gradual increase in OD values with experimental duration, while cells transfected with miR520a-3p mimics revealed a gradual decrease in OD values during the experimental course (Figure 2B). Compared to NC cells, cells transfected with miR520a-3p revealed decreased numbers cell clones (Figure 2, C and D) and invading cells (Figure 2, E and F). Also, scratch assays showed that wound width produced by NC cells was narrower than by cells transfected with miRNA mimics (Figure 2, G and H).
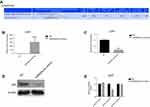
miR520a-3p directly targeted RELA/p65
Online software predicted that miR520a-3p would posttranscriptionally regulate RELA expression by hybridizing to a conserved site within RELA 3ʹUTR (Figure 3A), indicating that miR520a-3p can function in glioblastoma cells through suppressing RELA. In the present study, expression correlations of miR520a-3p with RELA were firstly analyzed through a gain-of-function approach. Endogenous RELA mRNA was detected in UC251 cells, and was significantly downregulated owing to transfection with miR520a-3p mimics (Figure 3, B and C). Also, transfection with miR520a-3p mimics resulted in barely detectable p65 protein in U251 cells (Figure 3D). Further, dual-luciferase reporter assays showed that RELA/p65 was a direct target of miR520a-3p (Figure 3E).
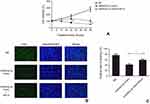
Restoration of RELA weakened the suppressor effect of miR520a-3p overexpression on proliferation of glioblastoma cells
Whether miR520a-3p functioned in glioblastoma by suppressing RELA was explored here by determining the effect of RELA restoration on miR520a-3p overexpression–induced inhibition of glioblastoma cells. During the 48-hour experimental course, transfection of miR520a-3p mimics led to a gradual decrease in viability, while cells displayed more viability at each time point owing to additional transfection of recombinant vectors containing RELA (Figure 4A). Similarly, overexpression of miR520a-3p generated a decrease in EdU-labeled cells, which was weakened significantly due to additional overexpression of RELA (Figure 4, B and C).
Discussion
miRNAs are small endogenous noncoding RNA molecules that can incorporate into an RNA-induced silencing complex (RISC) and recognizes target mRNAs through imperfect bases, commonly resulting in translation inhibition or destabilization of target mRNA. As such, miRNAs function as posttranscriptional regulators of gene expression, thereby influencing diverse biological activities and pathological conditions, including cancers. Indeed, various miRNAs hold potential as attractive biomarkers and/or therapeutic targets, due to their important roles during development and progression of malignant tumors.13
miR520a-3p is a mature miRNA produced from a transcript of the MIR520A gene, located on human chromosome 19q13.42. Various studies have documented that miR520a-3p is able to exert a suppressor role in the tumorigenesis of several malignant tumors, including non-small-cell lung cancer,14–16 colorectal cancer,17,18 thyroid carcinoma,19 osteosarcoma,20 and breast cancer.21 We found that miR520a-3p was downregulated in glioma tissue and downregulation of miR520a-3p associated with malignancy, suggesting that miR520a-3p should be a suppressor miRNA in glioma carcinogenesis. The ROC curve of miR520a-3p expression between normal brain tissue and gliomas showed that area under the curve was 0.9483, indicating that miR520a-3p expression was helpful in auxiliary glioma diagnosis. In this study, the suppressor role of miR520a-3p in glioma was validated through an in vitro approach. The data showed that miR520a-3p was able to suppress proliferation, migration, and invasion of glioblastoma cells.
miR520a-3p can bind the 3ʹUTR of different genes, including MAP3K2, HOXD8,14–16 EGFR,17,18 AKT1,19,20 CCD1, and CD44.21Online algorithms predicted conserved base pairing of RELA 3ʹUTR (position 1268–1275) with miR520a-3p, meaning that miR520a-3p exerted its role through targeting RELA. We found that miR520a-3p expression was correlated with RELA mRNA levels in glioma. In addition, transfection of miR520a-3p mimic actually resulted in downregulation of RELA/p65 in both mRNA and protein levels in glioblastoma cells. Dual-luciferase reporter assays showed that RELA/p65 was a direct target of miR520a-3p. Restoration of RELA significantly weakened the suppressor effect of miR520a-3p overexpression on proliferation of glioblastoma cells. Taken together, these findings suggest that miR520a-3p participates in the development and progression of glioblastoma through suppressing RELA. Attenuation in RELA/p65 may weaken the heterodimerization with p50, and thus impair the canonical NFκB signaling that has been considered an important participator in regulating development and progression of various types of solid tumors.
Conclusion
To our knowledge, this is the first report that miR520a-3p can inhibit growth of glioblastoma cells through suppressing RELA/p65. As such, miR520a-3p could be an attractive therapeutic target to eliminate glioblastoma, but complete understanding of the molecular mechanisms underlying the role of miR520a-3p in the development and progression of glioblastoma is still needed before clinical translation.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wesseling P, Capper D. WHO 2016 classification of gliomas. Neuropathol Appl Neurobiol. 2018;44(2):139–150.
2. Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–1850. doi:10.1001/jama.2013.280319
3. Zhang AS, Ostrom QT, Kruchko C, Rogers L, Peereboom DM, Barnholtz-Sloan JS. Complete prevalence of malignant primary brain tumors registry data in the United States compared with other common cancers, 2010. Neuro Oncol. 2017;19(5):726–735.
4. Hombach-Klonisch S, Mehrpour M, Shojaei S, et al. Glioblastoma and chemoresistance to alkylating agents: involvement of apoptosis, autophagy, and unfolded protein response. Pharmacol Ther. 2018;184:13–41.
5. Kim JS, Kim EJ, Kim HS, Kurie JM, Ahn YH. MKK4 activates non-canonical NFκB signaling by promoting NFκB2-p100 processing. Biochem Biophys Res Commun. 2017;491(2):337–342.
6. Xing Y, Wang X, Jameson SC, Hogquist KA. Late stages of T cell maturation in the thymus involve NF-κB and tonic type I interferon signaling. Nat Immunol. 2016;17(5):565–573.
7. Li Y, Zhou QL, Sun W, et al. Non-canonical NF-κB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation. Nat Cell Biol. 2015;17(10):1327–1338.
8. Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1(4):a000034.
9. Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391(6665):410–413.
10. Xia L, Tan S, Zhou Y, et al. Role of the NFκB-signaling pathway in cancer. Onco Targets Ther. 2018;11:2063–2073.
11. Nogueira L, Ruiz-Ontanon P, Vazquez-Barquero A, Moris F, Fernandez-Luna JL. The NFκB pathway: a therapeutic target in glioblastoma. Oncotarget. 2011;2(8):646–653.
12. Kaltschmidt B, Greiner JFW, Kadhim HM, Kaltschmidt C. Subunit-specific role of NF-κB in cancer. Biomedicines. 2018;6(2):E44.
13. Wang S, Yin Y, Liu S. Roles of microRNAs during glioma tumorigenesis and progression. Histol Histopathol. 2019;34(3):213–222.
14. Yu J, Tan Q, Deng B, Fang C, Qi D, Wang R. The microRNA-520a-3p inhibits proliferation, apoptosis and metastasis by targeting MAP3K2 in non-small cell lung cancer. Am J Cancer Res. 2015;5(2):802–811.
15. Liu Y, Miao L, Ni R, et al. microRNA-520a-3p inhibits proliferation and cancer stem cell phenotype by targeting HOXD8 in non-small cell lung cancer. Oncol Rep. 2016;36(6):3529–3535.
16. Lv X, Li CY, Han P, Xu XY. MicroRNA-520a-3p inhibits cell growth and metastasis of non-small cell lung cancer through PI3K/AKT/mTOR signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22(8):2321–2327.
17. Zhang R, Liu R, Liu C, et al. A novel role for MiR-520a-3p in regulating EGFR expression in colorectal cancer. Cell Physiol Biochem. 2017;42(4):1559–1574.
18. Qu X, Yang L, Shi Q, Wang X, Wang D, Wu G. Lidocaine inhibits proliferation and induces apoptosis in colorectal cancer cells by upregulating mir-520a-3p and targeting EGFR. Pathol Res Pract. 2018;214(12):1974–1979. doi:10.1016/j.prp.2018.09.012
19. Bi CL, Zhang YQ, Li B, Guo M, Fu YL. MicroRNA-520a-3p suppresses epithelial-mesenchymal transition, invasion, and migration of papillary thyroid carcinoma cells via the JAK1-mediated JAK/STAT signaling pathway. J Cell Physiol. 2018. doi:10.1002/jcp.27199.
20. Wang X, Xu Y, Chen X, Xiao J. Dexmedetomidine inhibits osteosarcoma cell proliferation and migration, and promotes apoptosis by regulating miR-520a-3p. Oncol Res. 2018;26(3):495–502.
21. Li J, Wei J, Mei Z, et al. Suppressing role of miR-520a-3p in breast cancer through CCND1 and CD44. Am J Transl Res. 2017;9(1):146–154.
Supplementary material
 |
Table S1 Clinicopathological features of glioma patients enrolled in this study |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.