Back to Journals » Cancer Management and Research » Volume 11
MiR-935 Promotes Clear Cell Renal Cell Carcinoma Migration and Invasion by Targeting IREB2
Authors Liu F, Chen Y, Chen B, Liu C, Xing J
Received 25 September 2019
Accepted for publication 16 December 2019
Published 30 December 2019 Volume 2019:11 Pages 10891—10900
DOI https://doi.org/10.2147/CMAR.S232380
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Alexandra R. Fernandes
This paper has been retracted.
Fei Liu, 1, 2 Yuedong Chen, 2 Bin Chen, 2 Chunxiao Liu, 1 Jinchun Xing 2
1Department of Urology, Zhujiang Hospital of Southern Medical University, Guangzhou 510280, People’s Republic of China; 2Department of Urology, The First Affiliated Hospital of Xiamen University, Xiamen 361003, People’s Republic of China
Correspondence: Chunxiao Liu
Department of Urology, Zhujiang Hospital of Southern Medical University, Guangzhou 510280, People’s Republic of China
Email [email protected]
Jinchun Xing
Department of Urology, The First Affiliated Hospital of Xiamen University, No. 55 Zhenhai Road, Xiamen 361003, People’s Republic of China
Tel +86-138 0608 9889
Email [email protected]
Purpose: Clear cell renal cell carcinoma (ccRCC) has the highest rate of metastasis and invasion in RCC and is the third most common adult urinary malignancy. miRNA may serve a critical role in human cancer development and progression, has been confirmed to play a pivotal role in RCC cell invasion and migration. Since miR‑935 had been verified to be an oncogene or tumor suppressor in various cancers, the role of miR‑935 in RCC was unclear.
Methods: Real-time quantitative polymerase chain reaction (qRT-PCR) was used to verify miR-935 expression. CCK-8 assay, wound healing assay and transwell assay were used to investigate the cell proliferation, migration and invasion of miR-935. Receiver operating characteristic (ROC) curve analysis was applied to discriminate different clinical classifications. Gain or loss of function approaches were used to investigate the cell proliferation, migration and invasion of miR-935 in vitro. Bioinformatics analysis and dual-luciferase reporter assay were used to identify the target of miR-935.
Results: MiR-935 had a higher expression level in RCC cells and cancer tissues. MiR-935 mimics promoted cell proliferation, migration and invasion, and miR-935 inhibitor inhibited cell inhibit malignancy of cancer cells. Bioinformatics analysis and dual-luciferase reporter assay identified iron-responsive element-binding protein 2 (IREB2) as a direct target of miR-935. qRT-PCR showed IREB2 expression was downregulated in ccRCC cancer tissues and high IREB2 expression had a longer overall survival (OS) and disease-free survival (DFS). Silencing IREB2 could reverse the function of miR-935 inhibitor on cell proliferation and metastasis in renal cancer cells.
Conclusion: The study indicated that miR-935 may act as an oncomiRNA and influenced migration and invasion progress of ccRCC by targeting IREB2. Oncogene miR-935 may be a molecular marker and uncover new strategies for ccRCC.
Keywords: miR-935, clear cell renal cell carcinoma, IREB2, migration, invasion
Introduction
Cancer is an important public health problem. Renal cell carcinoma (RCC) constitutes for more than 3% of all adult malignancies, is the most lethal urological malignancy with about 65,340 new cases and 14,970 deaths estimated for 2018 in the United States.1 Clear cell renal cell carcinoma (ccRCC), which has the highest rate of mortality, invasion and metastasis, is the most common RCC histological subtype.2 Prevention and detection of cancers at early stage can get better treatment outcomes and different situations affect renal cell carcinoma diagnosis. One-third of the patient’s present metastasis when they were primarily diagnosed with RCC in consequence of the inexistence of diagnostic biomarkers.3,4 More and more researchers focus on tumor diagnosis or prognosis biomarkers. It is meaningful to look for the timely diagnostic markers which may be involved in the occurrence or development of cancer thus improving the prognosis of ccRCC patients.
MiRNAs are widely accepted to play critical roles in the progression and metastasis of tumors, such as angiogenesis,5 chemosensitivity,6–8 cell differentiation and proliferation,9 tumor invasion and metastasis10,11 and apoptosis12,13. Furthermore, miRNAs may function as “oncogenes” (oncomiRs) or “tumor suppressor genes” in various cancers. Abnormal miRNA expression found in many cancers and abnormal miRNA expression reveals some correlation between tumor type and stage and miRNA expression.14 About half of miRNAs are on the side of the tumor-associated genome and suggest that miRNAs may play an important role in tumor progression.15 Mature miRNA regulates gene expression negatively by repressing the translation of the target gene’s proteins from messenger RNAs (mRNAs) or by binding to the 3ʹ-untranslated regions to increase the degradation of mRNAs.16,17 Recently, miR-935 is observed to be upregulated in pancreatic cancer,18 liver cancer19 and gastric cancer.20 However, the expression and potential roles of miR-935 in renal cancer progression are largely unknown.
In the present study, we explored the potential roles of the miR-935 in ccRCC and found that miR-935 was elevated in renal cancer cells and cancer tissues. MiR-935 promoted cell proliferation and invasion by targeting iron response element-binding protein 2 (IREB2). Altogether, our findings suggest that miR-935 as an oncogene and may provide new insights into the treatment of ccRCC.
Materials and Methods
Human Samples and Ethics Statement
Twenty-five patient samples with ccRCC were obtained from the Department of Urology, Zhujiang Hospital of Southern Medical University, Guangzhou, China between 2016 and 2018. Cancer tissues and normal kidney tissues were frozen in liquid nitrogen freshly after sample collection and then stored at −80 °C. Written, informed consent was obtained from each individual patient. The study was approved by the Institutional Review Board of Southern Medical University, in line with the Helsinki Declaration.
RNA Extraction and qRT-PCR
Tissue and cell RNA was extracted with the TRizol reagent (Thermo, Massachusetts, USA) as previous research.21 The RNA solution concentration and purity were measured with NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, USA) and then reverse transcription with 1 μg RNA. miRNA reverse transcription was accomplished by the RevertAid First-Strand cDNA Synthesis Kit (Thermo, Massachusetts, USA) and reverse transcript primers were obtained from RiboBio (RiboBio, Guangzhou, China). SYBR Green mix (Thermo, Massachusetts, USA) was performed for qRT-PCR analysis. Primers of mir-935 (MQPS0002274-1-100) and U6 (MQPS0000002-1-100) were purchased from RiboBio (RiboBio, Guangzhou, China) and gene primers of GAPDH and IREB2 were obtained from GENEWIZ (GENEWIZ, Suzhou, China). Samples were normalized to U6 and GAPDH, respectively. The relative expression of miR-935 was analyzed with the 2−ΔΔCt.
IREB2 (forward, 5ʹ-GCGATTTCCAGGCTTGCTTA-3ʹ;
reverse, 5ʹ-GTTTAACACGCAGACCAGCT −3ʹ).
GAPDH (forward, 5ʹ-GAGTCAACGGATTTGGTCGT-3ʹ;
reverse, 5ʹ-GACAAGCTTCCCGTTCTCAG-3ʹ)
Cell Culture and miRNA Mimics, Inhibitors and IREB2 siRNA Transfection
Human normal kidney cell (HK2), RCC cell lines 786-O, A498 and ACHN were obtained from the American Type Culture Collection (ATCC). Cells were cultured in DMEM medium with 10% FBS and 1% penicillin–streptomycin in 5% CO2 at 37°C. Cancer cells were seeded in six-well plates at a density of 1×105/well. miR-935 inhibitor, miRNA-935 mimic, IREB2 siRNA or their negative control were transfected into 780-O and A498 cells with Lipofectamine 2000 reagents (Thermo Fisher Scientific, Waltham, USA). RNA oligonucleotides were obtained from RiboBio (Guangzhou, China) as previous research.22 Forty-eight hours later, the cells were prepared for further analysis.
Cell Proliferation Assay
Cells (4×103) were seeded in 96-well plates for cell proliferation assay. Detection of cell growth rate was detected with the cell counting kit-8 (CCK-8) method according to the manufacturer’s instructions. Cell viability was assessed at 24, 48, 72 and 96 hrs cells were seeded, respectively.
Wound Healing Assay
Six-well plates were used for cell seed at a density of 1 ×105 cells per well. After transfection overnight, scratching the monolayer of cells with a sterile 10 ul micropipette tip. Washing with PBS for three times and then incubated cells in serum free medium at 37°C, 5% CO2. Observing and photographing the scratch healing area of cells at 0 h and 24 h.
Cell Migratory and Invasion Assays
Cell migratory and invasion were evaluated by transwell assay. Fifty-milliliter Matrigel (BD Biosciences, San Jose, CA, USA) was coated the upper chambers at 4°C. Cells were incubated without serum for 12 h, washed and resuspended with serum-free BSA (Invitrogen, NY, USA). The lower chamber was placed with 500 mL of DMEM high glucose medium (Invitrogen, NY, USA) containing 20% FBS (Invitrogen, NY, USA) as a chemokine. After 24-hr incubation, the upper chamber cells were removed and then fixed the chambers with 4% paraformaldehyde, washed the chambers and stained with 0.1% crystal violet (Thermo ScientificTM, #R40052, Waltham, MA, USA). Counting the migrated or invasion cells under the microscope. Three independent experiments were done.
Luciferase Assays
Reporter plasmids of wild-type or mutant IREB2 3ʹUTR were purchased from RiboBio (RiboBio, Guangzhou, China). Cells were transfected with 500ng luciferase reporter and cotransfected with miR-NC or miR-935 mimics in 24-well plates by Lipofectamine 2000 reagents (Thermo Fisher Scientific, Waltham, USA). The dual luciferase result is determined by the dual luciferase system assay (Promega, Madison, USA). Renilla-luciferase values were normalized to control reporter according to the manufacturer’s protocol.
Western Blotting
Cells are pyrolyzed in RIPA, protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA) and PMSF (Wuhan Boster Biological Technology, Ltd., Wuhan, China) protein lysis system. A total of 30 μg proteins were added to the SDS-PAGE gel system, then separated the proteins and transferred them to polyvinylidene fluoride (PVDF) membrane (EMD Millipore, Bedford, MA, USA) within 90 mins. After protein transfer to the PVDF membrane, the membrane was blocked in 5% skim milk within 1 hr. After cleaning the membrane with PBS 3 times, incubated the membrane with antibody against GAPDH (1:2000; BM3876; Wuhan Boster Biological Technology, Ltd., Wuhan, China) or IREB2 (1:1000; 23829-1-AP; Proteintech, Rosemont, USA) at 4°C overnight. After incubated the membranes 12–16 hrs, the membranes were washed and incubated the membranes for 2 h at room temperature with secondary antibodies (1:5000; BA1020; Wuhan Boster Biological Technology, Ltd., Wuhan, China). Finally, the membranes were detected by Biosense SC8108 Gel Documentation System with GeneScope V1.73 software (Shanghai BioTech, Shanghai, China) as previous research.23
Statistical Analysis
SPSS 22.0 software (SPSS, Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad, Software, San Diego, CA, USA) were used to data analyses and perform. The data were presented as the means ± standard deviation. Regarding statistical analysis, a normality test was performed on the expression levels of genes, p = 0.200, and the data conformed to a normal distribution. The correlation between miR-935 expression of patients and the clinicopathological parameters with ccRCC was evaluated with χ2 test. Student’s t-test was used to assess the differences of miR-935 expression between each ccRCC cancer and normal kidney tissues. The prognostic value of miR-935 for various ccRCC clinicopathological factors was evaluated using receiver operator characteristic (ROC) curve analysis. A P value of <0.05 was considered statistically significant.
Results
MiR-935 Is Elevated in ccRCC Cancer Tissues and Cell Lines
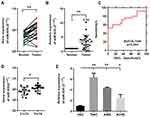
We used qRT-PCR to explore the expression of miR-935 in 25 paired ccRCC tissues and 3 RCC cell lines. Relationship between miR-935 expression and clinicopathological parameters is shown in Table 1. Compared with normal tissues, cancer tissues expressed higher levels of miR-935 (Figure 1A and B). ROC curve analysis showed that miR-935 could sufficiently discriminate ccRCC from normal tissues with an area under the curve (AUC) of 0.734 (95% CI: 0.5881 to 0.8807; P < 0.05) (Figure 1C). Moreover, we found a significantly higher miR-935 expression in T stage IV and III, when compared with T stage I and II (Figure 1D). We further investigate the expression level of miR-935 in normal kidney cell (HK2) and RCC cell lines (786-O, ACHN and A498). RCC cell lines expressed miR-935 highly when compared to HK2 (Figure 1D). Overall, these results indicate that miR-935 is overexpressed in RCC cell lines and tissues, suggesting that miR-935 may be a potential prognostic biomarker for ccRCC.
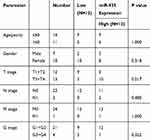 |
Table 1 Correlation Between miR-935 Expression and Clinicopathological Parameters of ccRCC Patients |
MiR-935 Promotes RCC Proliferation, Migration and Invasion in Renal Cancer Cells
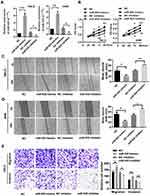
We selected the 786-O and A498 cell lines to evaluate the function of miR-935 in RCC as it is more highly expressed in those cell lines. We transfected NC, miR-935 inhibitor, or NC, miR-935 mimics into cells to investigate the influence of miR-935 on RCC cell growth. CCK-8 assay was conducted to investigate the influence of miR-935 on RCC cell growth, migration and invasion. The qRT-PCR results showed that the relative expression levels of miR-935 with miR-935 mimic and inhibitor-transfected 786-O and A498 cells are shown in Figure 2A. The results as shown in Figure 2B indicate that miR-935 inhibitors significantly impaired the proliferation and viability of 786-O and A498 cells. Transfection of miR‑935 mimic enhanced cell proliferation and viability of 786-O and A498 cells, suggesting that miR‑935 had an oncogenic effect. Wound healing experiments show that miR-935 inhibitors can significantly reduce the migration of 786-O and A498 cells, as shown in Figure 2C. As shown in Figure 2D, miR-935 mimics can enhance the migration of 786-O and A498 cells. Transwell analysis showed that miR-935 inhibitors inhibited the migration and invasion of 786-O cells, while miR-935 mimicked enhanced migration and invasion, as shown in Figure 2E.
IREB2 Is a Direct Target of miR-935
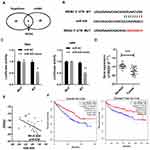
Two prediction software (TargetScan and miRDB) was used to predict the possible potential targets of miR-935. IREB2 was predicted to be a potential target of miR-935 as shown in Figure 3A. The dual-luciferase reporter assays were conducted to further validate whether miR-935 bound to the 3ʹ-UTRs of IREB2. Luciferase reporter constructs containing either the wild type (WT) or mutated (MUT) IREB2 binding sequences downstream of the firefly luciferase gene were generated (Figure 3B). Reporter vector plasmid and miR-935 mimics or mimics control were co-transfected into 786-O and A498 cells, luciferase activity was decreased significantly after miR-935 mimics co-transfection with WT vector plasmid (Figure 3C). These results imply that IREB2 was a direct target gene of miR-935. Then, we found IREB2 expression was lower in samples from ccRCC patients, and miR-935 expression was significantly negatively correlated with IREB2 in samples from ccRCC patients (Figure 3D and E). At last, we found high IREB2 expression had a longer survival on patient overall survival (OS) and disease-free survival (DFS) in renal cell carcinoma database from The Cancer Genome Atlas (TCGA) of ccRCC (TCGA_KIRC) (Figure 3F).
Silencing IREB2 Reverses the Function of miR-935 Inhibitor in Renal Cancer Cells
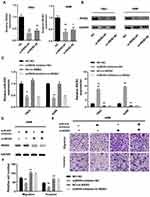
To find out whether IREB2 could reverse the function of miR-935, we co-transfected miR-935 inhibitor or/and siRNA of IREB2 into renal cancer cells. Two IREB2 siRNA were transfected into the A498 and 780-O cells (Figure 4A and B). The mRNA levels of miR-935 and IREB2 showed in Figure 4C, the protein levels of IREB2 were shown in Figure 4D. As shown in Figure 4E, knockdown IREB2 reversed the function of miR-935 inhibitor on the migration and invasion ability of A498 cells. Taken together, these data demonstrated that miR-935 could promote ccRCC cell migration and invasion by targeting IREB2.
Discussion
It is reported that miRNA is involved in tumorigenesis, acting variously as either oncogenes24–27 or tumor suppressors.16,28,29 Increasing evidence has demonstrated that miRNAs are effective biomarkers and tumor regulators in kidney cancer and therefore have broad implications in both clinical and therapeutic practice. Lin et al demonstrated that miR-154-5p regulates cell function and serves as a molecular marker for poor prognosis in renal cell carcinoma.30 Yu et al confirmed that miRNA-34a inhibits cell proliferation and metastasis in renal cancer cells by targeting CD44.31
As for miR-935, Wang et al found that miR-935 was up-regulated in pancreatic carcinoma and targeted inositol polyphosphate 4-phosphatase type I to promote cancer malignant behavior.18 Liu et al confirmed the truth that miR-935 could target SOX7 to promote liver cancer cell proliferation and migration.19 Yang et al informed that miR-935 promoted cell proliferation by targeting SOX7 in gastric cancer.20 Peng et al reported that miR-935 inhibition could increase paclitaxel sensitivity to non-small cell lung cancer via regulation of SOX7.32 Wang et al verified that mir-935 could downregulate IL-27 expression to inhibit suppression role of IL-27 in non-small-cell lung cancer cell.33 Huang et al illustrated that microRNA-935 was a prognostic marker and promoted cancer cell proliferation, migration, and invasion in colorectal cancer.34 In the microenvironment of solid tumors, intermediate-sized hyaluronan fragments could interact with TLR4 and then educate macrophage polarization to an M2-like phenotype via miR-935.35
In our study, dual-luciferase reporter assay and bioinformatics analysis identified iron-responsive element-binding protein 2 (IREB2) as a direct target of miR-935. Zhang et al revealed that miR-29 could bind on IREB2 and the expression of miR-29 was inversely correlated with IREB2 expression.36,37 IREB2 encodes a master regulator of iron metabolism including ferroptosis.38 Ferroptosis can inhibit the development of certain types of cancer, such as pancreatic cancer, hepatocellular carcinoma, breast cancer and prostate cancer.39,40 The role of IREB2 in kidney cancer has not been studied yet.
We discovered that miR-935 expression was upregulated in renal cancer tissues and cells. miR-935 could discriminate effectively between ccRCC and paired normal kidney tissues (AUC 0.7344; P<0.001) with ROC curve analysis. Then, we discovered patients who had renal carcinomas with high miR-935 expressions in T stage with low miR-935 expressions. This result might suggest miR-935 can be a potential diagnostic biomarker for renal cancer.
With the above results, we took further steps to evaluate the functional role of miR-935 downregulation or upregulation in renal cancer development. Through miR-935 inhibitor or mimics transduction, we showed that miR-935 had an oncogenic role by increasing proliferation, migration, and invasion in renal cancer cells. Downregulation of miR-935 had an anticancer effect in renal cancer cells. Dual-luciferase reporter assay and bioinformatics analysis identified that IREB2 as a direct target of miR-935. qRT-PCR showed IREB2 expression was downregulated in cancer tissues and high IREB2 expression had a longer OS and DFS. Silencing IREB2 could reverse the function of miR-935 inhibitor in renal cancer cell proliferation and metastasis.
Conclusion
Our results provided the first convincing evidence that miR-935-IREB2 may be an important oncogene by targeting IREB2 in human renal cancer. However, it might be limited in our research, and further research may be crucial for our future research. Whether miR-935 and IREB2 affect ferroptosis needs further study.
Data Sharing Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.
Funding
The present study was supported by scientific research grants from the Science and Technology Planning Project of the Guangzhou (201504301009390).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
2. Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi:10.1038/nrdp.2017.9
3. Linehan WM. The genetic basis of kidney cancer: implications for management and use of targeted therapeutic approaches. Eur Urol. 2012;61(5):896–898. doi:10.1016/j.eururo.2012.02.022
4. Bhatt JR, Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat Rev Urol. 2014;11(9):517–525. doi:10.1038/nrurol.2014.194
5. Sruthi TV, Edatt L, Raji GR, et al. Horizontal transfer of miR-23a from hypoxic tumor cell colonies can induce angiogenesis. J Cell Physiol. 2018;233(4):3498–3514. doi:10.1002/jcp.v233.4
6. Xiao W, Lou N, Ruan H, et al. Mir-144-3p promotes cell proliferation, metastasis, sunitinib resistance in clear cell renal cell carcinoma by downregulating ARID1A. Cell Physiol Biochem. 2017;43(6):2420–2433. doi:10.1159/000484395
7. Yue D, Qin X. miR-182 regulates trastuzumab resistance by targeting MET in breast cancer cells. Cancer Gene Ther. 2019;26(1–2):1–10. doi:10.1038/s41417-018-0031-4
8. Chen M, Wu L, Tu J, et al. miR-590-5p suppresses hepatocellular carcinoma chemoresistance by targeting YAP1 expression. EBioMedicine. 2018;35:142–154. doi:10.1016/j.ebiom.2018.08.010
9. Wang C, Su K, Zhang Y, et al. MicroRNA-365 targets multiple oncogenes to inhibit proliferation, invasion, and self-renewal of aggressive endometrial cancer cells. Cancer Manag Res. 2018;10:5171–5185. doi:10.2147/CMAR.S174889
10. Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi:10.1038/nature06174
11. Liu SY, Deng SY, He YB, Ni GX. miR-451 inhibits cell growth, migration and angiogenesis in human osteosarcoma via down-regulating IL 6R. Biochem Biophys Res Commun. 2017;482(4):987–993. doi:10.1016/j.bbrc.2016.11.145
12. Sharifi M, Moridnia A. Apoptosis-inducing and antiproliferative effect by inhibition of miR-182-5p through the regulation of CASP9 expression in human breast cancer. Cancer Gene Ther. 2017;24(2):75–82. doi:10.1038/cgt.2016.79
13. Dong X, Jin Z, Chen Y, et al. Knockdown of long non-coding RNA ANRIL inhibits proliferation, migration, and invasion but promotes apoptosis of human glioma cells by upregulation of miR-34a. J Cell Biochem. 2018;119(3):2708–2718. doi:10.1002/jcb.26437
14. Grady WM, Tewari M. The next thing in prognostic molecular markers: microRNA signatures of cancer. Gut. 2010;59(6):706–708. doi:10.1136/gut.2009.200022
15. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery. 2017;16(3):203–222. doi:10.1038/nrd.2016.246
16. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
17. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi:10.1038/nrm3838
18. Wang C, Feng Z, Jiang K, Zuo X. Upregulation of MicroRNA-935 promotes the malignant behaviors of pancreatic carcinoma PANC-1 cells via targeting inositol polyphosphate 4-phosphatase Type I gene (INPP4A). Oncol Res. 2017;25(4):559–569. doi:10.3727/096504016X14759554689565
19. Liu X, Li J, Yu Z, Li J, Sun R, Kan Q. miR-935 promotes liver cancer cell proliferation and migration by targeting SOX7. Oncol Res. 2017;25(3):427–435. doi:10.3727/096504016X14747300207374
20. Yang M, Cui G, Ding M, et al. miR-935 promotes gastric cancer cell proliferation by targeting SOX7. Biomed Pharmacother. 2016;79:153–158. doi:10.1016/j.biopha.2016.01.011
21. Xiao W, Wang X, Wang T, Xing J. TRIM2 downregulation in clear cell renal cell carcinoma affects cell proliferation, migration, and invasion and predicts poor patients’ survival. Cancer Manag Res. 2018;10:5951–5964. doi:10.2147/CMAR.S185270
22. Xiong Z, Xiao W, Bao L, et al. Tumor cell “Slimming” regulates tumor progression through PLCL1/UCP1‐mediated lipid browning. Adv Sci. 2019;1801862. doi:10.1002/advs.201801862
23. Xiao W, Xiong Z, Xiong W, et al. Melatonin/PGC1A/UCP1 promotes tumor slimming and represses tumor progression by initiating autophagy and lipid browning. J Pineal Res. 2019. doi:10.1111/jpi.12607
24. Li Q, Li B, Li Q, et al. Exosomal miR-21-5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition. Cell Death Dis. 2018;9(9):854. doi:10.1038/s41419-018-0928-8
25. Wen J, Hu Y, Liu Q, et al. miR-424 coordinates multilayered regulation of cell cycle progression to promote esophageal squamous cell carcinoma cell proliferation. EBioMedicine. 2018;37:110–124. doi:10.1016/j.ebiom.2018.10.043
26. Liu Y, Niu Z, Lin X, Tian Y. MiR-216b increases cisplatin sensitivity in ovarian cancer cells by targeting PARP1. Cancer Gene Ther. 2017;24(5):208–214. doi:10.1038/cgt.2017.6
27. Xiao W, Wang X, Wang T, Xing J. MiR-223-3p promotes cell proliferation and metastasis by downregulating SLC4A4 in clear cell renal cell carcinoma. Aging. 2019;11(2):615–633. doi:10.18632/aging.v11i2
28. Xu S, Tao Z, Hai B, et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun. 2016;7:11406. doi:10.1038/ncomms11406
29. Li YY, Tao YW, Gao S, et al. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine. 2018;36:209–220. doi:10.1016/j.ebiom.2018.09.006
30. Lin C, Li Z, Chen P, et al. Oncogene miR-154-5p regulates cellular function and acts as a molecular marker with poor prognosis in renal cell carcinoma. Life Sci. 2018;209:481–489. doi:10.1016/j.lfs.2018.08.044
31. Yu G, Li H, Wang J, et al. miRNA-34a suppresses cell proliferation and metastasis by targeting CD44 in human renal carcinoma cells. J Urol. 2014;192(4):1229–1237. doi:10.1016/j.juro.2014.05.094
32. Peng B, Li C, Cai P, Yu L, Zhao B, Chen G. Knockdown of miR935 increases paclitaxel sensitivity via regulation of SOX7 in non-small-cell lung cancer. Mol Med Rep. 2018;18(3):3397–3402. doi:10.3892/mmr.2018.9330
33. Wang T, Chen Y, Nie H, Huang Y, Zhao Y, Yang J. IL-27 inhibits non-small-cell lung cancer cell metastasis by miR-935 in vitro. Onco Targets Ther. 2019;12:1447–1454. doi:10.2147/OTT.S173207
34. Huang Y, Xiao W, Jiang X, Li H. MicroRNA-935 acts as a prognostic marker and promotes cell proliferation, migration, and invasion in colorectal cancer. Cancer Biomarkers. 2019;26(2):229–237. doi:10.3233/CBM-190183
35. Zhang B, Du Y, He Y, et al. INT-HA induces M2-like macrophage differentiation of human monocytes via TLR4-miR-935 pathway. Cancer Immunol Immunother. 2019;68(2):189–200. doi:10.1007/s00262-018-2261-6
36. Ripa R, Dolfi L, Terrigno M, et al. MicroRNA miR-29 controls a compensatory response to limit neuronal iron accumulation during adult life and aging. BMC Biol. 2017;15(1):9. doi:10.1186/s12915-017-0354-x
37. Zhang L, Ye Y, Tu H, et al. MicroRNA-related genetic variants in iron regulatory genes, dietary iron intake, microRNAs and lung cancer risk. Ann Oncol. 2017;28(5):1124–1129. doi:10.1093/annonc/mdx046
38. Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi:10.1016/j.cell.2012.03.042
39. Tang M, Chen Z, Wu D, Chen L. Ferritinophagy/ferroptosis: iron-related newcomers in human diseases. J Cell Physiol. 2018;233(12):9179–9190. doi:10.1002/jcp.v233.12
40. Saint-Germain E, Mignacca L, Vernier M, Bobbala D, Ilangumaran S, Ferbeyre G. SOCS1 regulates senescence and ferroptosis by modulating the expression of p53 target genes. Aging. 2017;9(10):2137–2162. doi:10.18632/aging.v9i10
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.