Back to Journals » OncoTargets and Therapy » Volume 10
miR-509-5p inhibits cellular proliferation and migration via targeting MDM2 in pancreatic cancer cells
Authors Li X, Li Y, Wan L, Chen R, Chen F
Received 15 December 2016
Accepted for publication 9 June 2017
Published 11 September 2017 Volume 2017:10 Pages 4455—4464
DOI https://doi.org/10.2147/OTT.S130378
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Chiung-Kuei Huang
Xin Li,1 Yan Li,2 Li Wan,3 Rui Chen,4 Fei Chen5
1Department of Pharmacology, School of Pharmaceutical Sciences, Guangzhou Medical University, Guangzhou, 2Department of Ophthalmology and Otorhinolaryngology, Lecong Hospital of Guangzhou Medical University, Foshan, 3Department of Laboratory Medicine, The Fourth Affiliated Hospital of Guangzhou Medical University, 4Key Laboratory for Major Obstetric Diseases in Guangdong Province, Key Laboratory of Reproduction and Genetics in Guangdong Higher Education Institutes, The Third Affiliated Hospital of Guangzhou Medical University, 5Department of Histology and Embryology, School of Basic Sciences, Guangzhou Medical University, Guangzhou, People’s Republic of China
Objective: This study aimed to explore the effect of miR-509-5p on pancreatic cancer progression and clarify the underlying mechanisms.
Methods: Real-time quantitative reverse transcription polymerase chain reaction was employed to determine miR-509-5p expression in pancreatic cancer tissues and noncancerous adjacent tissues. CCK-8 and Transwell experiments were employed to examine cellular proliferation, migration, and invasion after miR-509-5p mimic or inhibitor transfection. Bioinformatics tools were used to identify the target gene of miR-509-5p, and cotransfection of the target gene and miR-509-5p mimic was performed to determine the effect on the proliferation and migration of pancreatic cancer cells. A xenograft mouse model and histological analysis were also used to test the effect of miR-509-5p on tumor growth in vivo.
Results: miR-509-5p expression was dramatically downregulated in pancreatic cancer tissues and in pancreatic cancer cell lines. miR-509-5p mimic markedly inhibited PANC-1 cell proliferation, migration, and invasion. Conversely, miR-509-5p inhibitor promoted PANC-1 cell proliferation, migration, and invasion. Furthermore, the 3'UTR–specific target site luciferase reporter assay also showed that miR-509-5p negatively regulated MDM2 at the post-transcriptional level. miR-509-5p effectively reversed the MDM2 overexpression-induced increase in PANC-1 cell proliferation and invasion. Moreover, miR-509-5p inhibited tumor growth and accelerated cell death in the tumor samples.
Conclusions: Our results suggested that miR-509-5p served as a new tumor suppressor via targeting the MDM2 gene, inhibiting pancreatic cancer progression.
Keywords: miR-509-5p, pancreatic cancer, MDM2, proliferation, migration, invasion
Introduction
Pancreatic cancer is one of the most common types of cancer in the world. The median survival of this devastating malignancy is 3–6 months, and the 5-year survival rate is less than 5%.1–3 Despite improvements in surgical and adjuvant medical care, these figures have remained unchanged for more than 40 years, with the mortality close to the incidence. Therefore, there is great clinical significance in further elucidating the molecular mechanisms involving metastasis in pancreatic cancer and identifying novel markers for the diagnosis, prognosis, and treatment of pancreatic cancer patients.
Small noncoding RNAs (microRNAs, miRNAs) are ~22 nucleotides in length and regulate the expression of their target mRNA via translation inhibition or mRNA cleavage.4 miRNAs play a crucial role in biological development and differentiation.5 The dysregulation of miRNAs can also lead to cancer development and progression.4,5 Aberrant expression or gene mutations of miRNAs have been thoroughly studied in many types of tumors, including lymphoma, lung cancer, pancreatic cancer, leukemia, breast cancer, colon cancer, and liver cancer.6 However, the roles of miRNAs in pancreatic cancer remain largely unknown. miR-509-5p was previously shown to inhibit cervical cancer and hepatocellular carcinoma cell proliferation and invasion.6 However, the effects of miR-509-5p on pancreatic cancer progression and the molecular mechanisms involved remain unclear.
In this study, we determined the expression of miR-509-5p in pancreatic cancer tissues and cell lines and investigated the effects of miR-509-5p on the functions of pancreatic cancer cells as well as its regulation of the target gene MDM2. Our results showed that miR-509-5p expression was significantly downregulated in pancreatic cancer tissues and cell lines, suggesting that miR-509-5p may serve as a cancer suppressor via targeting the MDM2 gene.
Materials and methods
Sample collection
Human pancreatic cancer tissues (n=10) and matched noncancerous adjacent tissues (n=10) were collected in our hospital from June 2013 to May 2014. The tissues were preserved in liquid nitrogen for RNA extraction. To isolate islet cells, the pancreas was digested with collagenase, and tissue fragments were elutriated and then dissociated by enzymatic processing with DNase and trypsin (Gibco, Life Technologies, USA), separating the islet cells into single cells. All subjects provided written informed consent. The Institutional Animal Care and Use Committee (IACUC) of Guangzhou Medical University approved this research, and all experiments were conducted according to their approved protocols.
Cell culture
Human pancreatic cancer AsPC-1 and BxPC-3 cell lines (ATCC, Manassas, VA, USA) were cultured in RPMI1640 (Gibco, Life Technologies, USA), whereas pancreatic cancer PANC-1 cell, HEK293T, and isolated islet cells were cultured in DMEM (Gibco, Life Technologies, USA) supplemented with 10% FBS (Gibco, Life Technologies, USA) in 37°C incubator containing 5% carbon dioxide.
Real-time quantitative reverse transcription polymerase chain reaction
According to the manufacturer’s instructions, TRIzol Reagent (Invitrogen, USA) was used to isolate total RNA from tissue samples and cells. cDNA was synthesized using a miScript II RT first-strand synthesis kit (Qiagen, Germany), and real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) detection was performed using an miScript SYBR Green PCR Kit (Qiagen, Germany) according to the manufacturer’s protocols. The qRT-PCR amplification conditions for miR-509-5p consisted of 95°C for 15 min, followed by 40 cycles of 94°C for 15 s, 55°C for 30 s, and 70°C for 30 s. Human U6 RNA was used as a control to normalize miR-509-5p expression, and the ΔΔCt method was used to analyze the data as described previously.7
Western blotting
Cells were lysed in RIPA buffer supplemented with a protease inhibitor cocktail, and the bicinchoninic acid assay was used to quantify the protein concentrations. The proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the separated proteins were transferred to polyvinylidene difluoride membranes (Millipore). After blocking, the membranes were immunostained with mouse anti-MDM2 primary antibody (Santa Cruz Biotechnology, CA, USA) overnight, and then, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibody at room temperature followed by SuperSignal Ultra Chemiluminescent Substrate detection (Pierce, IL, USA). GAPDH (Santa Cruz Biotechnology) was used as a control to confirm equal sample loading.
Cell transfection
The PANC-1 cells were seeded in six-well plates before transfection and were cultured in DMEM for overnight. The cells were transfected with miR-509-5p mimic, inhibitor, or scramble control (Shanghai Genepharma Co., Ltd, Shanghai, People’s Republic of China) using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Cell proliferation
To evaluate cell proliferation, 1×104 PANC-1 cells were seeded in 96-well plates before transfection and cultured in DMEM containing 10% FBS overnight. The cells were transfected with miR-509-5p mimic, inhibitor, or scramble control or cotransfected with the target gene and miR-509-5p mimic or scramble control. After transfection for 24, 48, and 96 h, a CCK-8 (Dojindo, Kumamoto, Japan) was used to determine the cell proliferation with a microplate reader (Biotek ELX-800, USA) according to the manufacturer’s instructions.
Transwell migration and invasion assay
The cell migration and invasion assays were performed using the Transwell system. For migration assays, resuspended 5×104 PANC-1 cells were seeded into the upper chamber of the insert (BD Bioscience, 8-μm pore size). For invasion assays, 2×105 PANC-1 cells transfected with miR-509-5p mimic or inhibitor were seeded into the upper chamber coated with Matrigel, and the lower chamber was filled with DMEM containing 10% FBS. After being maintained in a cell culture incubator for a certain period, the membranes of the upper chamber were stained by hematoxylin, and three random fields (400× magnification) of each membrane were picked for cell number counting under a light microscope. All experiments were performed in triplicate.
Luciferase reporter assay
HEK293T cells were seeded in 96-well plates. When the cell growth reached 70% confluence, the cells were cotransfected with a Renilla luciferase reporter vector (20 ng), pLuc-3′UTR reporter vector (100 ng), negative control (5 pmol), and miR-509-5p mimic using Lipofectamine 2000. At 48 h after transfection, the Dual-Luciferase Reporter System (Promega, Madison, WI, USA) was used to measure the firefly and Renilla luciferase activities. The transfection efficiency was quantitated by normalization with a Renilla reporter vector.
Xenograft model
Six- to eight-week-old athymic male nude mice (SLAC Laboratories, Shanghai, People’s Republic of China) were maintained under a standard 12 h light/12 h dark cycle with food and water available ad libitum. The nude mice were inoculated subcutaneously with PANC-1 cells expressing miR-509-5p or the scramble control (5×106 cells each mouse, n=6–8). The tumor sizes of each mouse were monitored once a week. Tumor volumes were calculated as V =0.5 × D × d2, where D and d represent the larger and the smaller diameter of the xenograft tumor, respectively. When the tumor size reached ~1 cm in diameter, the mice were killed, and the xenograft tumors from each group were weighed. The dissected tumors were fixed in neutral buffered formalin and embedded in paraffin, and sections (5 μm) were prepared for histological examination. All the mouse experiments were performed under a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of Guangzhou Medical University.
Immunohistochemistry and histological staining
For the immunohistochemistry assay, the sections were deparaffinized and rehydrated in xylene, 99% ethanol, 96% ethanol, and 70% ethanol and then washed in distilled H2O. Antigen retrieval was carried out by boiling the samples in citrate buffer for 15 min at 92°C–98°C and trypsinizing with 0.125% trypsin (Invitrogen, New York, NY, USA) for 15 min at 37°C. The sections were immersed in 3% hydrogen peroxide and distilled water for 30 min to block endogenous peroxidase activity. Nonspecific staining was eliminated by a 20-min incubation with normal sheep serum (Invitrogen, New York, NY, USA). The sections were incubated with mouse anti-MDM2 antibody (1:200 dilution) at 37°C for 2 h and then incubated with goat anti-mouse secondary antibodies (IgG/HRP) (Zhongshan Goldenbridge Biotechnology Corp., Beijing, People’s Republic of China) for 15 min at 37°C using Non-Bio Two-Step Histostain™-Plus kits with DAB staining (Zhongshan Goldenbridge Biotechnology Corp.). The nuclei were counterstained with hematoxylin. The remaining procedures were performed in accordance with the manufacturer’s instructions.
For histological examination, the sections were stained with hematoxylin and eosin (HE). Both immunohistochemically stained and HE-stained sections were observed by light microscopy (Olympus Optical Co., Lake Success, NY, USA).
Statistical analyses
Data were analyzed and presented as the mean ± SD using SPSS 18 (SPSS, Chicago, IL, USA). The significant difference was examined with Student’s t-test or one-way analysis of variance.
Results
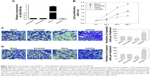
miR-509-5p is downregulated in pancreatic cancer tissues and cell lines
The expression levels of miR-509-5p in pancreatic cancer tissues, noncancerous adjacent normal tissues, isolated islet cells, and three pancreatic cancer cell lines were examined by qRT-PCR. Compared to the adjacent normal tissues, the level of miR-509-5p was significantly downregulated in pancreatic cancer tissues (Figure 1). Furthermore, we found a remarkably downregulated level of miR-509-5p in pancreatic cancer AsPC-1, BxPC-3, and PANC-1 cell lines compared with isolated islet cells (Figure S1).
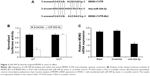
miR-509-5p inhibits cellular proliferation, migration, and invasion in pancreatic cancer cells
As the PANC-1 cell line exhibited the lowest level of miR-509-5p among three cancer cell lines, according to Figure S1, we used the PANC-1 cell line in the subsequent experiments and performed miR-509-5p mimic and inhibitor transfection (Figure 2A). miR-509-5p upregulation could significantly inhibit cell proliferation, whereas miR-509-5p downregulation could significantly increase cellular proliferation (Figure 2B). The miR-509-5p mimic remarkably inhibited the cellular migration and invasion of PANC-1 cells, whereas the miR-509-5p inhibitor exerted a reverse effect on PANC-1 cell migration and invasion (Figure 2C and D).
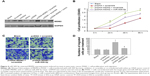
miR-509-5p targets MDM2
The TargetScan, PicTar, and miRanda software programs were used to predict miRNA targets. According to the predicted results, we found that the putative target gene of miR-509-5p is MDM2 (Figure 3A). Then, we constructed the luciferase vectors MDM2-WT and MDM2-Mut to validate the direct target gene. Compared to the scramble control, a decreased level of luciferase activity was found in cells transfected with MDM2-WT vector and miR-509-5p (Figure 3B). We also examined the effect of miR-509-5p on the regulation of MDM2. Compared to the scramble control group, PANC-1 cells treated with miR-509-5p mimic showed decreased MDM2 mRNA levels (Figure 3C).
miR-509-5p reverses MDM2 overexpression-increased cellular proliferation and migration in pancreatic cancer cells
To further verify MDM2 expression, we constructed MDM2-overexpressing PANC-1 cells and performed cell proliferation and migration analyses.
First, we examined the effect of miR-509-5p on the regulation of the MDM2 protein level. Compared to the miR-509-5p scramble control group, miR-509-5p mimic decreased the MDM2 protein level, but miR-509-5p inhibitor increased the MDM2 protein level (Figure 4A). Additionally, compared to the scramble control group, the PANC-1 cells overexpressing MDM2 showed an increased MDM2 protein level, while miR-509-5p mimic decreased the MDM2 protein level (Figure 4A). Moreover, MDM2 overexpression was found to significantly increase PANC-1 cell proliferation, while miR-509-5p mimic could reverse this effect exerted by MDM2 (Figure 4B). The cell migration results showed that MDM2 overexpression significantly increased PANC-1 cell migration, while miR-509-5p mimic could inhibit the MDM2-induced increase in cell migration (Figure 4C and D).
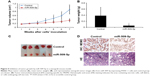
Inhibition of tumor growth by miR-509-5p in a xenograft mouse model
Finally, we tested the effect of miR-509-5p in a xenograft mouse model. Nude mice were subcutaneously inoculated with human PANC-1 cells expressing miR-509-5p or the scramble control. The volume of tumors treated with miR-509-5p decreased at a higher rate than in the control group (Figure 5A). Moreover, the weight of tumors treated with miR-509-5p showed a marked decrease compared with the control group (Figure 5B). The tumor images were consistent with the tumor weights (Figure 5C). As shown in Figure 5D, the relative expression intensity of MDM2-positive cells in tumors treated with miR-509-5p was lower than in the control group (Figure 5D, upper left and right panels). Histological analysis revealed that tumors from the scramble control group only contained small necrotic areas occasionally (Figure 5D, lower left panel). In contrast, tumors treated with miR-509-5p contained large areas filled with necrotic cell debris and enlarged necrotic cores (Figure 5D, lower right panel), which suggested that miR-509-5p inhibited tumor growth and accelerated cell death in the tumor samples.
Discussion
miRNAs play an important role as regulators of gene expression at post-transcriptional levels and regulate a wide range of physiological and developmental processes.8–10 There is increasing evidence that changes in the expression of miRNAs result in the pathogenesis of most human cancers and that they may act as either oncogenes or tumor suppressors.11,12 Recently, accumulated data suggest that miRNAs are involved in several important biological events, such as tumorigenesis and cancer metastasis.13,14
In this study, we investigated for the first time the biological role of miR-509-5p in human pancreatic cancer progression. Our results demonstrated that miR-509-5p was frequently downregulated in human pancreatic cancer tissues and pancreatic cancer cell lines. Enhanced miR-509-5p expression could inhibit the cellular proliferation, migration, and invasion of pancreatic cancer in vitro and tumor growth in vivo. We further identified the direct functional target of miR-509-5p, MDM2, a putative oncogene. Our results show that miR-509-5p acts as a new tumor suppressor gene in pancreatic cancer.
There is evidence that miR-509-5p can suppress the cell growth capacity of human cervical cancer and hepatocellular carcinoma.15 However, the expression of miR-509-5p in pancreatic cancer and its function in pancreatic cancer progression remain unclear. In this study, we determined the expression of miR-509-5p in clinical pancreatic cancer tissues, noncancerous adjacent tissues, and several pancreatic cancer cell lines by qRT-PCR. The results show that the expression of miR-509-5p was downregulated compared with the matched paired noncancerous adjacent tissues, indicating that it may be involved in both carcinogenic transformation and tumor metastasis. Furthermore, the expression of miR-509-5p was remarkably decreased in all three pancreatic cancer cell lines compared with isolated islet cells. Subsequently, we found that the downregulation or upregulation of miR-509-5p significantly promoted or inhibited, respectively, the cellular proliferation, migration, and invasion of pancreatic cancer cells in vitro.
Bioinformatics approaches were used to predict the target gene of miR-509-5p. We further explored the underlying molecular mechanism involved and identified MDM2 as a direct functional target of miR-509-5p in pancreatic cancer cells. A well-conserved miR-509-5p-binding site was identified in the 3′UTR of MDM2 mRNA, and we demonstrated that miR-509-5p directly targets the 3′UTR of MDM2, as its enhanced expression suppressed luciferase activity. In addition, miR-509-5p overexpression significantly decreased the expression of MDM2 protein, indicating the post-transcriptional regulation of MDM2 via targeting its 3′UTR. These results suggested that the function of miR-509-5p as a tumor suppressor may be mediated via repressing MDM2 expression in pancreatic cancer progression.
MDM2, as an oncogene, was first found in a locus amplified on double minute chromosomes in a tumorigenic mouse cell line (3T3-DM).16 MDM2 mainly inhibits p53 bioactivity by blocking its transcriptional activity and promoting its degradation,17,18 decreasing p53 protein levels and attenuating p53 function, which increases cancer risk and accelerates tumor formation and progression.19 The MDM2 oncogene plays a key role in cancer progression.20 MDM2 overexpression could induce tumor cell proliferation and inhibit cell apoptosis.21
Several studies have shown that MDM2 overexpression is associated with poor survival and is a useful predictive factor for poor prognosis in patients with hepatocellular carcinoma and breast cancer.22,23 A recent study reported that the expression level of MDM2 was much higher in pancreatic cancer tissues than in paired normal pancreatic tissues, which was associated with clinicopathological features in patients treated only with surgery.24 Moreover, MDM2 knockdown has a synergistic effect on the suppression of pancreatic cancer cell growth in vivo.25 However, the mechanism by which MDM2 is regulated remains unknown. Our experimental results indicated that the role of miR-509-5p targeting MDM2 may provide one such mechanism for the post-transcriptional regulation of MDM2. In addition, we observed the effects of miR-509-5p overexpression on a tumor xenograft model. It was noteworthy that, in agreement with the outcomes obtained in vitro, the overexpression of miR-509-5p mimics in vivo resulted in decreased tumor growth, accompanied by the acceleration of cell death.
Currently, the emergence of new technologies that use synthetic miRNA mimics or anti-miRNA oligonucleotides exhibits great promise in clinical miRNA therapy,26 whereas no clinical application of synthetic miRNA mimics has yet been reported. Synthetic miR-509-5p mimic treatment in cancer will become a significant scientific and therapeutic challenge. Although there is increasing evidence that miRNAs might help to generate targeted therapies and to overcome conventional anticancer resistance, the development of a safe and efficient method for clinical use remains one of the main hurdles of miRNA-based therapy. Of course, it has to be acknowledged at this stage that the translation of these results from cells/mice to clinical application is not feasible. Furthermore, the therapeutic delivery of miRNAs is still a developing field, and much more work needs to be done before miRNAs can be safely applied in clinical practice. However, these findings provide a very promising basis for future studies to determine the effect of miRNA modulation on chemotherapy in clinical application.
In summary, our results have shown that miR-509-5p is significantly downregulated in pancreatic cancer tissues and cell lines. The enhanced expression of miR-509-5p decreased pancreatic cancer cell proliferation, migration, and invasion by directly targeting MDM2. These findings suggest that this novel miR-509-5p/MDM2 axis provides new insight into the mechanisms underlying tumor metastasis and that the repression of miR-509-5p expression may be a potential therapeutic strategy for the treatment of pancreatic cancer in the future.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31401240, 81202604), Guangdong Natural Science Foundation (2015A030310153), Guangdong Medical Research Foundation (B2014185, B2014190), and Guangzhou Medical University Science Foundation (2013C01).
Disclosure
The authors report no conflicts of interest in this work.
References
Lin QJ, Yang F, Jin C, Fu DL. Current status and progress of pancreatic cancer in China. World J Gastroenterol. 2015;21(26):7988–8003. | ||
He Y, Zheng R, Li D, Zeng H, Zhang S, Chen W. Pancreatic cancer incidence and mortality patterns in China, 2011. Chin J Cancer Res. 2015;27(1):29–37. | ||
Chen W, Zheng R, Zhang S, et al. Annual report on status of cancer in China, 2010. Chin J Cancer Res. 2014;26(1):48–58. | ||
Li Z, Lei H, Luo M, et al. DNA methylation downregulated mir-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer. 2015;18(1):43–54. | ||
Wang HJ, Ruan HJ, He XJ, et al. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur J Cancer. 2010;46(12):2295–2303. | ||
Yu X, Zhang X, Bi T, et al. MiRNA expression signature for potentially predicting the prognosis of ovarian serous carcinoma. Tumour Biol. 2013;34(6):3501–3508. | ||
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. | ||
Furuta M, Kozaki K, Tanimoto K, et al. The tumor-suppressive miR-497–195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013;8(3):e60155. | ||
Namlos HM, Meza-Zepeda LA, Baroy T, et al. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7(10):e48086. | ||
Zhao G, Cai C, Yang T, et al. MicroRNA-221 induces cell survival and cisplatin resistance through PI3K/Akt pathway in human osteosarcoma. PLoS One. 2013;8(1):e53906. | ||
Kobayashi E, Hornicek FJ, Duan Z. MicroRNA involvement in osteosarcoma. Sarcoma. 2012;2012:359739. | ||
Truini A, Coco S, Alama A, et al. Role of microRNAs in malignant mesothelioma. Cell Mol Life Sci. 2014;71(15):2865–2878. | ||
Yongchun Z, Linwei T, Xicai W, et al. MicroRNA-195 inhibits non-small cell lung cancer cell proliferation, migration and invasion by targeting MYB. Cancer Lett. 2014;347(1):65–74. | ||
Craig VJ, Cogliatti SB, Rehrauer H, Wundisch T, Muller A. Epigenetic silencing of microRNA-203 dysregulates ABL1 expression and drives Helicobacter-associated gastric lymphomagenesis. Cancer Res. 2011;71(10):3616–3624. | ||
Ren ZJ, Nong XY, Lv YR, et al. Mir-509-5p joins the Mdm2/p53 feedback loop and regulates cancer cell growth. Cell Death Dis. 2014;5:e1387. | ||
Pichiorri F, Suh SS, Rocci A, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010;18(4):367–381. | ||
Zhao Y, Yu H, Hu W. The regulation of MDM2 oncogene and its impact on human cancers. Acta Biochim Biophys Sin. 2014;46(3):180–189. | ||
Nag S, Zhang X, Srivenugopal KS, Wang MH, Wang W, Zhang R. Targeting MDM2-p53 interaction for cancer therapy: are we there yet? Curr Med Chem. 2014;21(5):553–574. | ||
Nag S, Qin J, Srivenugopal KS, Wang M, Zhang R. The MDM2-p53 pathway revisited. J Biom Res. 2013;27(4):254–271. | ||
Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. | ||
Li Q, Lozano G. Molecular pathways: targeting Mdm2 and Mdm4 in cancer therapy. Clin Cancer Res. 2013;19(1):34–41. | ||
Peng Q, Lao X, Chen Z, et al. TP53 and MDM2 gene polymorphisms, gene-gene interaction, and hepatocellular carcinoma risk: evidence from an updated meta-analysis. PLoS One. 2013;8(12):e82773. | ||
Park HS, Park JM, Park S, Cho J, Kim SI, Park BW. Subcellular localization of Mdm2 expression and prognosis of breast cancer. Int J Clin Oncol. 2014;19(5):842–851. | ||
Sheng W, Dong M, Zhou J, et al. The clinicopathological significance and relationship of Gli1, MDM2 and p53 expression in resectable pancreatic cancer. Histopathology. 2014;64(4):523–535. | ||
Wang W, Qin JJ, Voruganti S, et al. Identification of a new class of MDM2 inhibitor that inhibits growth of orthotopic pancreatic tumors in mice. Gastroenterology. 2014;147(4):893–902.e892. | ||
Garofalo M, Croce CM. microRNAs: master regulators as potential therapeutics in cancer. Annu Rev Pharmacol Toxicol. 2011;51:25–43. |
Supplementary material
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.