Back to Journals » OncoTargets and Therapy » Volume 9
MiR-502-3P suppresses cell proliferation, migration, and invasion in hepatocellular carcinoma by targeting SET
Authors Jin H, Yu M, Lin Y, Hou B, Wu Z, Li Z, Sun J
Received 23 April 2015
Accepted for publication 19 October 2015
Published 31 May 2016 Volume 2016:9 Pages 3281—3289
DOI https://doi.org/10.2147/OTT.S87183
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Faris Farassati
This paper has been retracted.
Haosheng Jin,* Min Yu,* Ye Lin, Baohua Hou, Zhongshi Wu, Zhide Li, Jian Sun
Department of General Surgery, Guangdong General Hospital, Guangdong Academy of Medical Sciences, Guangzhou, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Background/aim: Increasing evidences show that microRNAs are engaged in hepatocellular carcinoma (HCC). The aim of this study was to investigate the role of miR-502-3P in HCC and to identify its underlying mechanism.
Methods: The expression levels of miR-502-3P were assessed in multiple HCC cell lines and in liver tissues of patients with HCC. We further examined the effects of miR-502-3P on malignant behavior of HCC. The molecular target of miR-502-3P was identified using a computer algorithm and confirmed experimentally.
Results: Downregulation of miR-502-3P was found in both HCC cell lines and human samples. Overexpression of miR-502-3P dramatically inhibits HCC proliferation, metastasis, invasion, and cell adhesion. We further verify the SET as a novel and direct target of miR-502-3P in HCCs.
Conclusion: Taken together, overexpression of miR-502-3P or downregulation of SET may prove beneficial as a therapeutic strategy for HCC treatment.
Keywords: miRNA, HCC, SET gene, recurrence
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world, especially in East Asia.1–3 Approximately 60,000 people died of HCC each year, and it is now the second leading cause of cancer death worldwide. Only ~10%–30% of patients have the opportunity for surgery, which is mainly liver resection and liver transplantation.4 The prognosis of HCC is still dismal due to the late diagnosis and high rate of recurrence. Thus, further exploring the mechanisms underlying initiation, progression, and metastasis of HCC is helpful for early detection and effective treatment of HCC. MicroRNAs (miRNAs) are a class of small, short noncoding RNAs, which are proved to have dual roles in the development and progression of HCC. More and more evidence showed that miRNAs are able to act as oncogenes or tumor suppressor in various human cancers.5–7 Previously, we reported the role of miR-26b in modulating the epithelial–mesenchymal transition and its relationship with poor survival of HCC.8 As miRNA expression profilings are extensively used, many potential miRNAs that are involved in the development and progression of HCC are identified,9–12 such as miR-10a-5p, miR-122-5p, miR-146b-5p, miR-148a-3p, miR-26, miR-29, and miR-221.12,13 We previously also performed miRNA profilings and found that several miRNAs were significantly dysregulated in HCC, including miR-502-3P. Further analysis indicated that downexpression of miR-502-3P was associated with postoperative recurrence and Edmonson grade.14 However, the function of miR-502-3P is still unclear. In this study, we identified that a novel miRNA, miR-502-3P, was frequently downregulated in HCC cell lines and HCC tissues. We found that overexpression of miR-502-3P inhibited the proliferation, metastasis, invasion, and cell adhesion. We further identified the SET gene as a direct target of miR-502-3P in HCCs. Therefore, our data strongly suggested that miR-502-3P is a tumor suppressor by targeting SET expression to modulate HCC malignant biological behavior. Overexpression of miR-502-3P or downregulation of SET may be helpful for developing new strategies for HCC treatment.
Methods and materials
Patients’ selection
Histologically confirmed HCC samples were derived from 50 patients undergoing surgical resection at Guangdong General Hospital. All the patients signed the written consent forms indicating their willingness to participate in this study. This study complied with the Declaration of Helsinki and the use of human cell lines was approved by the Institutional Ethics Committee of Guangdong General Hospital. All the included pathologically and histologically confirmed patients with HCC met the following criteria: no history of any other malignant tumor, without any local or systemic anticancer treatment prior to the surgery. Samples were immediately snap frozen and stored in liquid nitrogen for RNA analysis. The tumor tissue was chosen from a region without necrosis or hemorrhaging, while the paratumor liver tissue was gathered within a 5 cm distance of the tumor.15
Cell culture
The following human HCC cell lines were included in this study: MHCC-97H, SMMC-7221, HepG2, Huh-7, and Hep3B. The normal hepatocyte LO2 was also employed as normal control. All the cell lines were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA).
Cell transfections
Transfection of the miR-502-3P mimics was performed using Lipofectamine™ RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instructions.
RNA extraction and real-time PCR analysis
Total miRNA from cultured cells and fresh surgical HCC tissues was extracted using TRIzol reagent (Thermo Fisher Scientific), and the concentration of the total RNA was quantitated by measuring the absorbance at 260 nm. Complementary DNA was generated using a miScript Reverse Transcription Kit (Qiagen NV, Venlo, the Netherlands). Primers for miR-502-3P and the U6 small nuclear RNA (snRNA, internal control) were purchased from Land (Guangzhou, Guangdong, People’s Republic of China). The expression level of miRNA was defined based on the threshold cycle (Ct), and relative expression levels were calculated using the 2−ΔΔCt method, using the expression level of the U6 snRNA as a reference gene. Each polymerase chain reaction (PCR) was performed in triplicate. The primers for the examined genes are presented in Table 1.
  | Table 1 Primer for qRT-PCR |
Cell proliferation
MTT assay (Hoffman-La Roche Ltd, Basel, Switzerland) was used to evaluate the HCC cell proliferation after treatments. In this assay, 2×103 cells were seeded into 96-well plates, and cell numbers were calculated using MTT assay according to the manufacturer’s instructions.
Invasion and migration assay
The invasion and migration assay was performed using transwell chambers consisting of 8 mm membrane filter inserts (Corning Incorporated, Corning, NY, USA). After being trypsinized and suspended in serum-free medium, 1.5×105 cells were seeded into transwell insert supplemented with RPMI-1640 with 10% serum. The bottom side of transwells was filled with RPMI-1640 with 20% serum. Matrigel (Becton-Dickinson, Mountain View, CA, USA) was seeded into transwell insert and allowed to grow to confluence for 1 day in invasion assay. After 36 hours incubation, cells that had invaded the lower chamber were fixed with 4% paraformaldehyde and stained with hematoxylin, and the number of cells on the lower side of the filter was counted under a microscope. Ten different views were randomly chosen, and the average count was taken. This experiment was performed independently three times in duplicates.
Wound healing assay
Cells were trypsinized and seeded in equal numbers into six-well tissue culture plates and allowed to grow until confluent (~24 hours). Following serum starvation for 24 hours, an artificial homogeneous wound (“scratch”) was created on- the cell monolayer with a sterile 100 μL tip. After scratching, the cells were washed with serum-free medium, complete media were added, and microscopic images (20× magnification) of the cells were collected at 0 hour, 12 hours, and 24 hours.
Cell adhesion assay
Cells were trypsinized and seeded in equal numbers to each of the collagen I-coated wells. The plate was incubated at 37°C for 20 minutes to allow the cells to adhere to the surface. After washing with DMEM, DMEM with 10% fetal bovine serum was added, and the cells were incubated at 37°C for 4 hours for recovery. Then, 10 μL of MTT substrate was added to each well, and incubation was continued for an additional 2 hours at 30°C. Next, the MTT-treated cells were lysed, and the absorbance at 570 nm was measured on a spectrophotometer.
Annexin V-fluorescein isothiocyanate and propidium iodide staining
Cells induced by various treatments were gently trypsinized and collected by centrifugation. After resuspension with binding buffer, cells were incubated in fluorescein isothiocyanate conjugated to annexin V/propidium iodide according to the manufacturer’s instructions. The population of annexin V–propidium iodide viable cells and annexin V + apoptotic cells was evaluated by flow cytometry. Data were collected in a FACS Calibur (Becton-Dickinson) and analyzed using Cell Quest software (Becton-Dickinson).
Luciferase reporter assay
Luciferase reporter assay was performed according to the manufacturer’s instructions. Briefly, cells (3.5×104) were seeded in triplicate in 24-well plates overnight. Next, 100 ng of pGL3-SET-3′UTR (wt/mut) or control-luciferase plasmid plus 1 ng of pRL-TK renilla plasmid (#E2810; Promega Corporation, Fitchburg, WI, USA) were transfected into the cells using Lipofectamine 2000 (Thermo Fisher Scientific). Three independent experiments were performed, and the data are presented as the mean ± standard deviation.
Statistical analysis
Student’s t test was used to evaluate the statistical significance of the difference between the two groups of continuous data, whereas chi-square test was applied for the analysis of categorical data. P-value of <0.05 was considered to be statistically significant. All analyses in this study were performed by SPSS 13.0 (SPSS Inc., Chicago, IL, USA) statistical software package.
Results
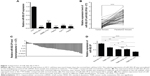
MiR-502-3P was downregulated in HCC cell lines and human tissues
To evaluate the role of miR-502-3P in HCC, we detected the expression level of miR-502-3P in HCC cell lines and human tissues from patients with HCC. The results showed that expression of miR-502-3P was obviously downregulated in HCC cell lines, including MHCC-97H, SMMC-7221, HepG2, Huh-7, and Hep3B, compared with normal live cell LO2 (Figure 1A). Detection of miR-502-3P was also performed in HCC tissues from patients, and results indicated that mirR-502-3P was significantly downregulated in tumor tissue compared with adjacent nontumor tissues (median =1.07 vs 2.83; P<0.001) (Figure 1B and C). We divided the included patients according to the tumor–node–metastasis (TNM) classification15,16 and found that the expression level of miR-502-3P was significantly higher in the HCC samples at early stages (TNM I and II) compared to that of the HCC samples at advanced tumor stages (TNM III) (Figure 1D). These results suggested that miR-502-3P expression was significantly and negatively associated with TNM stage in HCC.
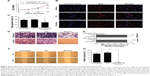
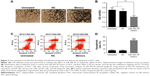
Overexpression of miR-502-3P inhibits the proliferation, metastasis, invasion, and cell adhesion properties but induces the apoptosis of HCC cells
To assess the role of miR-502-3P in HCC proliferation, we transfected miR-502-3P mimics into HepG2 cell. Result of MTT assay suggested that cells treated with miR-502-3P mimics showed a significantly slower rate than did control cells after culturing for 2 days (P<0.05 at day 2, P<0.01 at day 3) (Figure 2A). Furthermore, EdU(5-ethynyl- 2′-deoxyuridine) incorporation analysis of S-phase cells indicated that miR-502-3P mimics significantly inhibited S-phase entry and progression of cell cycle, compared to control (37.1% vs 58.2%, P<0.01) (Figure 2B and C). Transwell assay depicted that treatment with miR-502-3P mimics resulted in a significant decrease in the metastasis potential of HCC (7.3-fold reduction, P<0.01), whereas invasion assay indicated that treatment with miR-502-3P contributed to dramatic decrease in the invasive property of HCC (99.9-fold reduction, P<0.001) (Figure 2D and E). Wound healing assay showed that the mobility of HCC cells was evidently suppressed after treatment with miR-502-3P mimics, compared to control (wound closure, 20.1% vs 0.8%, P<0.001) (Figure 2F and G). Cell adhesion assay indicated that miR-502-3P mimics also remarkably inhibits cell adhesion compared with control (optical density value, 0.41 vs 0.63, P<0.01) (Figure 3A and B). We also detected the apoptotic population of HCC cells by flow cytometry after various treatments. The results showed that apoptotic populations of HCC cells induced by miR-502-3P mimics were 62.15%, compared with untreated group (P<0.001) and negative control of mimics (P<0.001) (Figure 3C and D). Our results generally showed that miR-502-3P can induce apoptosis in HCC cell lines.
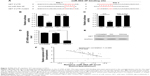
MiR-502-3P regulates SET expression in HCC
To determine the underlying mechanisms by which miR-502-3P regulates the malignant biological behavior of HCC, we integrated bioinformatics algorithms, including miRanda, PicTar, and TargetScan to predict the potential targets of miR-502-3P. By using bioinformatics algorithms, we assumed that SET was a putative target gene of miR-502-3P (Figure 4A). As shown in Figure 4B and C, luciferase reporters indicated that wild type 3′-untranslated region resulted in a significant reduction in luciferase activity, whereas mutations of the key binding region showed no variation compared with control. The quantitative real-time PCR analysis suggested that treatment with miR-502-3P significantly suppressed the mRNA and protein expression of SET (Figure 4D and E). To further validate the correlation between miR-502-3P and SET, we employed PCR to investigate the expression level of SET in included human HCC samples. As indicated in Figure 4F, results of spearman’s rank test showed an inverse correlation between miR-502-3P and SET expression (r=−0.689, P=0.009). In summary, our data strongly suggested that SET is a novel target gene of miR-502-3P.
Discussion
Emerging evidence indicated that miRNAs have dual functions as oncogenes or tumor suppressors in human cancers depending on their specific target genes.17 Hence, further validation of potential important miRNAs involved in initiation, progression, and metastasis provides valuable insight for the diagnosis and therapy of patients with HCC. As the development of modern technology, miRNA profilings are cheap, and feasible. As in HCC, miRNA profilings were carried out by a few researchers, and lots of valuable miRNAs were identified.18–20 Although tremendous efforts have been put into the investigation of miRNAs function, the roles of miRNAs in the molecular pathogenesis of HCC remain largely unknown.
We previously reported that expression of miR-502-3P was dramatically decreased in HCC with early recurrence (<1 year) compared to nonearly recurrence (>2 years), which suggested that miR-502-3P may play an important role in HCC pathogenesis and recurrence.11,16,17 However, the expression and the function of miR-502-3P in HCC remain obscure. Therefore, this study was carried out to investigate the role of miR-502-3P in HCC. We found that miR-502-3P was significantly downregulated in both HCC cell lines and human samples. Moreover, expression of miR-502-3P was inversely correlated with the severity and progression of HCC, which suggested that decreased expression of miR-502-3P may correlate with malignant biological behavior. Therefore, we further detected the expression of miR-502-3P on malignant behavior of HCC. Results manifested that overexpression of miR-502-3P significantly inhibited proliferation, metastasis, invasion, and cell adhesion of HCC in vitro. However, the underlying mechanism of tumor suppressor role of miR-502-3P needed further elucidation. By using the bioinformatics algorithms, SET gene may hold immense probability as target gene of miR-502-3P. Previous data have demonstrated that SET is a potent and specific inhibitor of protein phosphatase 2A and is associated with many cellular processes, such as cell cycle control,21 migration,22,23 and apoptosis.24 Previous reports showed that increased expression of SET was found in head and neck squamous cell carcinoma,25 breast cancer,26 leukemia,27 lung cancer,28 and pancreatic cancer.29 Of note, no reports about SET gene in HCC were published so far. As far as we know, this study was the first study concerned with SET gene in HCC, and the molecular mechanism is still unclear. To validate the SET gene as the target gene of miR-502-3P, luciferase reporter was employed, and we found that miR-502-3P remarkably suppressed the expression of SET gene. Moreover, enforced expression of miR-502-3P significantly reduces miRNA and protein level of SET in HCC cell line. We further validated the correlation of miR-502-3P and SET gene in human HCC samples. In summary, these results suggested that miR-502-3P has a tumor suppressor role in HCC, and it may exert its specific role by repressing the SET gene expression.
In conclusion, the results suggested that miR-502-3P inhibits the malignant behavior of HCC by, at least partly, repressing the expression of SET gene. Therefore, upregulation of miR-502-3P or suppression of SET gene may prove beneficial as a therapeutic strategy for HCC treatment.
Acknowledgment
This study was supported by a grant from the National Science Foundation of Guangdong Province, People’s Republic of China (No. 2014A030310073), Guangdong Provincial Science and Technology Plan projects (No. 2009B080701021 and 2010B080701021)Guangdong Province Public interest research and capacity - building projects, People’s Republic of China (No.2014A020212448) and Guangzhou Science and technology plan of scientific research projects, People’s Republic of China (No. 201510010286).
Disclosure
The authors report no conflicts of interest in this work.
References
Ganne-Carrie N, Mohand D, N’kontchou G, Grando-Lemaire V, Trinchet JC. [Synopsis: diagnosis and treatment of hepatocellular carcinoma in patients with cirrhosis]. Gastroenterol Clin Biol. 2002; 26(1):73–77. Chinese. | ||
Befeler AS, Di Bisceglie AM. Hepatocellular carcinoma: diagnosis and treatment. Gastroenterology. 2002;122(6):1609–1619. | ||
Chen MF. Icteric type hepatocellular carcinoma: clinical features, diagnosis and treatment. Chang Gung Med J. 2002;25(8):496–501. | ||
Lau WY, Lai EC. Hepatocellular carcinoma: current management and recent advances. Hepatobiliary Pancreat Dis Int. 2008;7(3):237–257. | ||
Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. | ||
Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353(17):1768–1771. | ||
Voorhoeve PM. MicroRNAs: oncogenes, tumor suppressors or master regulators of cancer heterogeneity? Biochim Biophys Acta. 2010;1805(1):72–86. | ||
Shen G, Lin Y, Yang X, Zhang J, Xu Z, Jia H. MicroRNA-26b inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting USP9X. BMC Cancer. 2014;14:393. | ||
Toffanin S, Hoshida Y, Lachenmayer A, et al. MicroRNA-based classification of hepatocellular carcinoma and oncogenic role of miR-517a. Gastroenterology. 2011;140(5):1618.e–1628.e. | ||
Li W, Lebrun DG, Li M. The expression and functions of microRNAs in pancreatic adenocarcinoma and hepatocellular carcinoma. Chin J Cancer. 2011;30(8):540–550. | ||
Wei L, Lian B, Zhang Y, et al. Application of microRNA and mRNA expression profiling on prognostic biomarker discovery for hepatocellular carcinoma. BMC Genomics. 2014;15(suppl 1):S13. | ||
Murakami Y, Tanahashi T, Okada R, et al. Comparison of hepatocellular carcinoma miRNA expression profiling as evaluated by next generation sequencing and microarray. PLoS One. 2014;9(9):e106314. | ||
Braconi C, Henry JC, Kogure T, Schmittgen T, Patel T. The role of microRNAs in human liver cancers. Semin Oncol. 2011;38(6):752–763. | ||
Zhi-liang D, Zhi-xiang J, Jian S. The expression and clinical significance of miR-144 and miR-502-3P in early recurrence of hepatocellular carcinoma. Re Dai Yi Xue Za Zhi. 2012;12(6):679–682. | ||
Liu Y, Ding Y, Huang J, et al. MiR-141 suppresses the migration and invasion of HCC cells by targeting Tiam1. PLoS One. 2014; 9(2):e88393. | ||
Tu H, Wei G, Cai Q, et al. MicroRNA-212 inhibits hepatocellular carcinoma cell proliferation and induces apoptosis by targeting FOXA1. Onco Targets Ther. 2015;8:2227–2235. | ||
Yang J, Han S, Huang W, et al. A meta-analysis of microRNA expression in liver cancer. PLoS One. 2014;9(12):e114533. | ||
Cairo S, Wang Y, de Reyniès A, et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci U S A. 2010;107(47):20471–20476. | ||
Chung GE, Yoon JH, Myung SJ, et al. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol Rep. 2010;23(1):113–119. | ||
Diaz G, Melis M, Tice A, et al. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. 2013;133(4):816–824. | ||
Canela N, Rodriguez-Vilarrupla A, Estanyol JM, et al. The SET protein regulates G2/M transition by modulating cyclin B-cyclin-dependent kinase 1 activity. J Biol Chem. 2003;278(2):1158–1164. | ||
ten Klooster JP, Leeuwen Iv, Scheres N, Anthony EC, Hordijk PL. Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. EMBO J. 2007;26(2):336–345. | ||
Lam BD, Anthony EC, Hordijk PL. Cytoplasmic targeting of the proto-oncogene SET promotes cell spreading and migration. FEBS Lett. 2013; 587(2):111–119. | ||
Madeira A, Pommet JM, Prochiantz A, Allinquant B. SET protein (TAF1beta, I2PP2A) is involved in neuronal apoptosis induced by an amyloid precursor protein cytoplasmic subdomain. FASEB J. 2005;19(13):1905–1907. | ||
Sobral LM, Sousa LO, Coletta RD, et al. Stable SET knockdown in head and neck squamous cell carcinoma promotes cell invasion and the mesenchymal-like phenotype in vitro, as well as necrosis, cisplatin sensitivity and lymph node metastasis in xenograft tumor models. Mol Cancer. 2014;13:32. | ||
Li J, Yang XF, Ren XH, et al. Stable SET knockdown in breast cell carcinoma inhibits cell migration and invasion. Biochem Biophys Res Commun. 2014;453(1):7–12. | ||
Liu Y, He P, Liu F, Cheng X, Zhang M. [Influence of I2PP2A gene silencing by RNA interference on proliferation and apoptosis of human acute promyelocytic leukemia cell line NB4-R1]. Zhonghua Xue Ye Xue Za Zhi. 2014;35(8):732–736. Chinese. | ||
Saddoughi SA, Gencer S, Peterson YK, et al. Sphingosine analogue drug FTY720 targets I2PP2A/SET and mediates lung tumour suppression via activation of PP2A-RIPK1-dependent necroptosis. EMBO Mol Med. 2013;5(1):105–121. | ||
Farrell AS, Allen-Petersen B, Daniel CJ, et al. Targeting inhibitors of the tumor suppressor PP2A for the treatment of pancreatic cancer. Mol Cancer Res. 2014;12(6):924–939. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.