Back to Journals » OncoTargets and Therapy » Volume 9
MiR-203 is involved in the laryngeal carcinoma pathogenesis via targeting VEGFA and Cox-2
Authors Xu L, Shen B, Chen T, Dong P
Received 9 September 2015
Accepted for publication 5 February 2016
Published 26 July 2016 Volume 2016:9 Pages 4629—4637
DOI https://doi.org/10.2147/OTT.S96053
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Lin Xu,1 Bin Shen,2 Tingting Chen,3 Pin Dong2
1Department of Otolaryngology, Second Affiliated Hospital of Zhejiang University Medical College, Hangzhou, 2Department of Otolaryngology-Head & Neck Surgery, Shanghai Jiaotong University Affiliated First People’s Hospital, Shanghai, 3Lishui Central Hospital, Lishui, Zhejiang Province, People’s Republic of China
Abstract: The development of laryngeal squamous cell carcinoma (LSCC) is a multistep process involving multiple factors. MicroRNAs, a group of important negative regulators of gene expression, have also been confirmed to be involved in the LSCC pathogenesis. In the present study, we compared the expression of nine selected microRNAs in the LSCC tissues and adjacent nontumor tissues. We found that the expression of miR-203 was significantly reduced in the LSCC tissues. Predicted by using bioinformatics tools, we found that VEGFA and cyclooxygenase-2 (Cox-2) may be direct targets of miR-203. By subsequent determination through dual-luciferase assay and Western blot, we confirmed that miR-203 suppresses the expression of VEGFA and Cox-2 by directly targeting 3'-untranslated region. Meanwhile, by analyzing the relationship between miR-203 and VEGFA in clinical tissue samples, we found that a negative correlation existed in the expression of miR-203 and VEGFA (P=0.0096, r=-0.33). Similarly, the expression of miR-203 and Cox-2 also has a negative correlation (P=0.0019, r=-0.46). Subsequently, in vitro functional study indicated that miR-203 played as a tumor suppressor by repressing proliferation, migration, and invasion of Hep-2 cells. The overexpression of VEGFA partially rescued the effect of overexpressed miR-203. Overexpressed Cox-2 partially rescued the effect of miR-203 on Hep-2 cell proliferation but not on the cell migration and invasion capacity. These findings suggest that miR-203 plays as a tumor suppressor in LSCC, partially by regulating VEGFA and Cox-2, and may serve as a potential target for therapeutic intervention.
Keywords: non-coding RNA, laryngeal squamous cell carcinoma, correlation analysis
Introduction
Head and neck cancer is the seventh most common form of cancer in the world, which is composed of oral cavity, oropharynx, hypopharynx, and larynx cancers.1,2 Laryngeal cancer is considered to be the most common malignant neoplasm in the head and neck region. In 2008, an estimate of 151,000 new cases of laryngeal cancer was diagnosed worldwide. In the same year, it was responsible for 82,000 deaths worldwide.2 Meanwhile, although early stage laryngeal cancer is often cured by surgery or radiotherapy, for the majority of patients with the advanced disease, the outcome has not improved in the last three decades. This implies that significant health benefits could be achieved via effective prevention and treatment strategies.
MicroRNA (miRNA) is a kind of short noncoding RNAs that suppress the expression of protein-coding genes by partial complementary binding, especially to the 3′-untranslated regions (UTRs) of messenger RNAs. Alterations in MiRNA expression are involved in the initiation, progression, and metastasis of human cancer, and it is believed that miRNAs function both as tumor suppressors and as oncogenes in cancer development.3,4 Accumulating studies have shown that disturbed expression of miRNAs is involved in the cancer pathogenesis and drug resistance.5,6 Recently, increased reports indicated that disturbed miRNA levels existed in the laryngeal cancer and played very important roles in the pathogenesis of laryngeal cancer.7–9 However, the functional study is at the early stage and almost all the results needed to be confirmed in larger populations. Therefore, in this study, we detected the expression of nine miRNAs in laryngeal cancer tissues and examined the function of miR-203 on cell proliferation, migration, and invasion.
Patients and methods
Tissue samples
Patients were enrolled in the period between July 2010 and June 2013. A total of 60 patients with laryngeal squamous cell carcinoma (LSCC) who underwent partial or total laryngectomy at the Department of Otolaryngology – Head & Neck Surgery, Shanghai Jiao Tong University Affiliated First People’s Hospital were included in the study. No chemotherapy and radiation were applied to the patients before the operation. The matched specimens of LSCC and the corresponding adjacent nonneoplastic tissues obtained from the patients were preserved in liquid nitrogen within 5 minutes of excision and then transported frozen to the laboratory and stored briefly at -80°C. The study was approved by the Ethics Committee of Shanghai Jiao Tong University Affiliated First People’s Hospital, and written informed consent was obtained from all the participants.
RNA isolation and quantitative reverse transcription-polymerase chain reaction
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis was used to determine the relative expression levels of selected miRNAs. Total RNA was extracted from the tissue samples by using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Random six RNA samples were mixed as one sample for the detection of target molecules as the preliminary experiment. The levels of miR-203 in the samples of every patient were detected subsequently. The miRNAs expression was detected by TaqMan miRNA RT-Real-Time PCR. Single-stranded complementary DNA was synthesized by using TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) and then amplified by using TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) together with miRNA-specific TaqMan MGB probes (Thermo Fisher Scientific). The U6 snRNA was used for normalization. Each sample in each group was measured in triplicate, and the experiment was repeated at least three times for the detection of miRNAs.
Cell culture
Hep-2 and HEK293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (HyClone, Logan, UT, USA), 100 IU/mL penicillin, and 10 mg/mL streptomycin. All cells were maintained at a temperature of 37°C under an atmosphere of 5% CO2.
Western blot
Protein extracts were boiled in sodium dodecyl sulfate/β-mercaptoethanol sample buffer, and 30 μg samples were loaded into each lane of 10% polyacrylamide gels. The proteins were separated by electrophoresis, and the proteins in the gels were blotted onto polyvinylidene difluoride membranes (Amersham Pharmacia Biotech, St Albans, Herts, UK) by electrophoretic transfer. The membranes were incubated with rabbit anti-vascular endothelial growth factor A VEGFA polyclonal antibody (ab51745; Abcam, Cambridge, MA, USA) with a dilution of 1:2,000, rabbit anti-Cox-2 polyclonal antibody (Abcam) with a dilution of 1:5,000, or mouse anti-β-actin monoclonal antibody (sc-47778; Santa Cruz Biotechnology Inc., Dallas, TX, USA) with a dilution of 1:3,000 for 2 hours at 37°C. The specific protein–antibody complex was detected by using horseradish peroxidase-conjugated goat antirabbit or rabbit antimouse IgG. Detection by the chemiluminescence reaction was carried out using the ECL kit (Pierce, Appleton, WI, USA). The β-actin signal was used as a loading control.
Dual-luciferase assay
A 435 bp segment of VEGFA 3′-UTR and a 1132 bp segment of Cox-2 were cloned into downstream of firefly luciferase coding region in the pGL3 control vector (Promega Corporation, Fitchburg, WI, USA) to generate luciferase reporter vector. For luciferase reporter assays, HEK293T cells were seeded in 48-well plates. One of the luciferase reporter vectors was cotransfected with miR-203 into HEK293T cells by using lipofectamine 2000 (Thermo Fisher Scientific). After 48 hours, the cells were harvested and assayed with the Dual-Luciferase Assay (Promega Corporation). Each treatment was performed in triplicate in three independent experiments. The results were expressed as relative luciferase activity (Firefly luciferase/Renilla luciferase).
Cell proliferation assay
Hep-2 cells were seeded in 96-well plates at low density (5×103) in DMEM culture and allowed to attach overnight. The cells were then transfected with miR-203 mimic or inhibitor, with sequence scrambled single-strand or double-strand short RNA as control. In all, 20 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; 5 mg/mL; Sigma-Aldrich Co., St Louis, MO, USA) was added into each well after 48 hours of transfection, and the cells were incubated for further 4 hours. The absorbance was recorded at A570 nm with a 96-well plate reader after the dimethyl sulfoxide addition.
Cell migration and invasion assays
Hep-2 cells were transfected with the miR-203 mimic or inhibitor with scramble RNA as control. The transfected cells were harvested and subjected to the following assays after 48 hours of transfection. For migration assays, the transfected cells (5×104 cells/mL) were seeded in the top of an 8.0 mm-pore membrane chamber (Corning Incorporated, Corning, NY, USA). Following a 12-hour incubation period, the cells that passed through the membrane to attach to the bottom of the membrane were fixed and stained with hematoxylin and eosin (Sigma-Aldrich Co.). The cells were scraped and removed from the top of the chamber. The membranes were mounted on cover slides, and cells were counted. Cell migration was quantified by counting the amount of cells passing through the pores from five different fields per sample at 100× selected in a random manner. Cell invasion assays were carried out using modified Boyden chambers in 24-well tissue culture plates at 5×104 cells/well (BD Biosciences, San Jose, CA, USA). All experiments were performed in duplicate.
Statistical analysis
The results of independent two groups were analyzed by using t-test, and the correlation analysis was analyzed by χ2 test. All the data were analyzed by using SPSS Statistical Package Version 16 (SPSS Inc., Chicago, IL, USA). P<0.05 was considered statistically significant.
Results
Expression of miR-203 is reduced in LSCC tissues
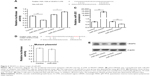
MiRNAs is a group of gene expression suppressors, and disturbed miRNAs expression has been found in the LSCC tissues. To investigate the roles of miRNAs in the pathogenesis of LSCC, we first selected nine miRNAs that were reported to have disturbed expression in the LSCC patients.7–9 Subsequently, total RNA was extracted from the LSCC tissues and adjacent nontumor tissues and every six RNA samples were mixed into one group. The expressions of miRNAs were detected by using stem-loop qRT-PCR. As shown in Figure 1, the expression of miR-203 was reduced significantly (P=0.0020) compared with the adjacent nontumor control. Since miR-203 has significant lower expression in tumor samples, the level of miR-203 in every tumor and adjacent nontumor samples was detected by qRT-PCR subsequently. The correlation of miR-203 and clinic pathological parameters was analyzed. Lower miR-203 expression was also found in the T3–T4 stage LSCC compared with T1–T2, and more reduced miR-203 expression was related to lymph node metastasis (Table 1).
  | Table 1 Correlation between miR-203 and clinicopathological parameters |
miR-203 suppresses the expression of VEGFA and Cox-2 by targeting 3′-UTR
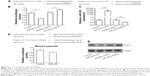
To understand the roles of reduced miR-203 in the pathogenesis of LSCC, we first predicted the target genes of miR-203 by using online bioinformatics tool: TargetScan (http://www.targetscan.org).10 To our surprise, VEGFA and Cox-2, the overexpression of whom has been found to be positively correlated with the malignancy of LSCC, are the direct feasible targets of miR-203 (Figures 2 and 3).10–13 To examine whether these two genes are the real target of miR-203, we employed the dual-luciferase reporter system. A 334 bp segment of VEGFA or 1138 bp segment of Cox-2 containing the putative target sites of miR-203 was subcloned into pGLO3 control vector, under the firefly luciferase coding region to generate the reporter plasmids. HEK293T cells were cotransfected with one of the reporter vectors and miR-203 mimics or antagonist, with pRL-TK which expression renilla luciferase as transfection control. The expression of miR-203 was detected by qRT-PCR (Figures 2B and 3B). Compared with the miRNA control, the luciferase activity was significantly suppressed by the miR-203, ~45.7% (P<0.01). Furthermore, the luciferase activity was significantly upregulated by the miR-203 inhibitor compared with the inhibitor control, ~12.5% (P<0.05). These results indicate that miR-203 targets the 3′-UTR of VEGFA, leading to the change in the translation of firefly luciferase. Similar results were obtained when using Cox-2 reporter vector. Compared with the miRNA control, the luciferase activity was significantly suppressed by the miR-203, ~44.1% (P<0.05). Furthermore, the luciferase activity was significantly upregulated by the miR-203 inhibitor compared with the inhibitor control, ~18.5% (P<0.05).
Seed sequence mutation clones were also used to further confirm the binding site for miR-203 (Figures 2D and 3D). The vector contains putative miR-203 binding region in the 3′-UTR of VEGFA with four mutant nucleotides or the 3′-UTR region of Cox-2 with six mutant nucleotides. The histogram in Figures 2D and 3D showed that the enzyme activity was not significantly reduced in cells transfected with miR-control compared with miR-203 mimic (P>0.05). These data indicate that miR-203 may suppress the expression of VEGFA and Cox-2 through binding to seed sequence at the 3′-UTR of VEGFA and Cox-2.
To further examine whether endogenous expression of VEGFA and Cox-2 is suppressed by miR-203, Hep-2 cells were transfected with miR-203 mimics or inhibitor. VEGFA and Cox-2 protein levels were detected by Western blot after 48 hours of transfection. Compared with the corresponding control, the levels of VEGFA and Cox-2 proteins were significantly suppressed by miR-203 mimics and upregulated by miR-203 inhibitor in Hep-2 cells (Figures 2E and 3E). These results indicated that miR-203 repressed the endogenous expression of VEGFA and Cox-2 in the laryngeal cancer cells by directly targeting 3′-UTR.
Expression of VEGFA and Cox-2 is negatively correlated with miR-203 level in vivo
To further examine the relationship between miR-203 level and its target genes in vivo, we detected the expression of miR-203 in every independent sample, and meanwhile, the protein levels of VEGFA and Cox-2 were also detected by Western blot. The relative band intensity was quantified by using ImageJ software (National Institutes of Health [NIH] Bethesda, MD, USA) with β-actin as the loading control. Analyzed with the expression of miR-203, VEGFA, and Cox-2, we found inverse correlations between the expression levels of miR-203 and VEGFA or Cox-2 in 60 clinical samples of LSCC. Low levels of miR-203 were associated with high expression of VEGFA (Pearson’s correlation, r=-0.398; P<0.01; Figure 4A and B) and Cox-2 (Pearson’s correlation, r=-0.302; P<0.05; Figure 4C and D).
miR-203 suppresses proliferation, migration, and invasion of laryngeal cancer cells
To further test whether miR-203 may execute tumor-suppressive functions by targeting VEGFA and Cox-2, we investigated the effect of miR-203-mediated cell proliferation by MTT assay in Hep-2 cells (Figure 5A). The cell proliferation was significantly reduced by miR-203 mimic, ~54.8%. Meanwhile, the cell proliferation was significantly upregulated by miR-203 inhibitor in Hep-2 cells, ~33.1%.
To further confirm whether miR-203 represses cell proliferation by targeting VEGFA and Cox-2, we cotransfected VEGFA or Cox-2 expression vector with miR-203 mimic into Hep-2 cells. As shown in Figure 5B, VEGFA and Cox-2 expression vector can partially rescue the repressed cell proliferation caused by miR-203 overexpression, which means that miR-203 suppresses the ability of cell proliferation partially through targeting VEGFA and Cox-2.
In order to further research the roles of miR-203 in controlling the metastasis of laryngeal cancer, we analyzed the effects of miR-203 on the migratory and invasive behaviors of Hep-2 cells. As shown in Figure 5C and E, the migration and invasion capacities were significantly suppressed in cells transfected with miR-203 mimic compared with control (P<0.05). In the contrary, the number of migrated and invaded cells was significantly upregulated by the miR-203 inhibitor (P<0.05). Meanwhile, VEGFA overexpression can partially rescue the repressed cell migration and invasion capacity caused by miR-203 (P<0.05) Figure 5D and F. Cox-2 overexpression can also slightly rescue the cell migration and invasion capacity, but the differences were not significant (P>0.05) Figure 5D and F. These results suggest that the level of miR-203 is closely associated with the metastasis of laryngeal cancer.
Discussion
The development of LSCC is a multistep process involving multiple factors. miRNAs, a group of important negative regulator of gene expression, have also been confirmed to be involved in the LSCC pathogenesis. In the present study, we compared the expression of nine selected miRNAs in the LSCC tissues and adjacent nontumor tissues. We found that the expression of miR-203 was significantly reduced in the LSCC tissues. Predicted by using bioinformatics tools, we found that VEGFA and Cox-2 might be the direct targets of miR-203. By subsequent determination through dual-luciferase assay and Western blot, we confirmed that miR-203 suppresses the expression of VEGFA and Cox-2 by directly targeting 3′-UTR. Meanwhile, by analyzing the relationship between miR-203 and VEGFA in clinical tissue samples, we found that a negative correlation existed in the expression of miR-203 and VEGFA. Similarly, the expression of miR-203 and Cox-2 also had a negative correlation. Subsequently, in vitro functional study indicated that miR-203 played as a tumor suppressor by repressing proliferation, migration, and invasion of Hep-2 cells. The overexpression of VEGFA partially rescued the effect of overexpressed miR-203. Overexpressed Cox-2 partially rescued the effect of miR-203 on the proliferation of Hep-2 cell but not on the cell migration and invasion capacity. These results indicated that miR-203 plays as a tumor suppressor in LSCC, partially by regulating VEGFA and Cox-2.
Although miR-203 has been reported to be upregulated in some kinds of tumors,5,14 it is widely accepted that miR-203 functions as a tumor suppressor and has been downregulated especially in head and neck cancer, prostate cancer, gastric cancer, and colon cancer.15–17 Attempts to validate miR-203 target genes have increasingly been made to investigate the functional role of miR-203 in carcinogenesis. In this study, we identified for the first time that VEGFA and Cox-2 are the direct targets of miR-203. The function of VEGFA is related to not only regulating vasculogenesis and angiogenesis but also affecting the tumor microenvironment. Cox-2 has prostaglandin (PG) hydroperoxidase activity to synthesize PGH2 from PGG2, and the expression of Cox-2 is induced during inflammation, cell proliferation, and differentiation. COX-2 levels have been found to be elevated in various tumors.18–20 In this study, we confirmed that negative correlations existed between the expression of miR-203 and VEGFA or Cox-2, which contributes to understanding the mechanism of the higher level of VEGFA and Cox-2 in the LSCC tissues.
Conclusion
miR-203 functions as a tumor suppressor via targeting VEGFA and Cox-2, and downregulated miR-203 expression contributes to LSCC carcinogenesis through enhanced tumor cell proliferation, migration, and invasion.
Disclosure
The authors report no conflicts of interest in this work.
References
Ahmad Kiadaliri A, Jarl J, Gavriilidis G, Gerdtham UG. Alcohol drinking cessation and the risk of laryngeal and pharyngeal cancers: a systematic review and meta-analysis. PLoS One. 2013;8(3):e58158. | ||
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. | ||
Wu WK, Lee CW, Cho CH, et al. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;29(43):5761–5771. | ||
Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs – the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9(4):293–302. | ||
Ikenaga N, Ohuchida K, Mizumoto K, et al. MicroRNA-203 expression as a new prognostic marker of pancreatic adenocarcinoma. Ann Surg Oncol. 2010;17(12):3120–3128. | ||
Izumchenko E, Chang X, Michailidi C, et al. The TGFbeta-miR200-Mig6 pathway orchestrates the EMT-associated kinase switch that induces resistance to EGFR inhibitors. Cancer Res. 2014;74(14):3995–4005. | ||
Hui AB, Lenarduzzi M, Krushel T, et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010;16(4):1129–1139. | ||
Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15(8):2850–2855. | ||
Zhang T, Liu M, Wang C, Lin C, Sun Y, Jin D. Down-regulation of MiR-206 promotes proliferation and invasion of laryngeal cancer by regulating VEGF expression. Anticancer Res. 2011;31(11):3859–3863. | ||
Agarwal V, Bell GW, Nam J, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife, 4:e05005. | ||
Sullu Y, Gun S, Atmaca S, Karagoz F, Kandemir B. Poor prognostic clinicopathologic features correlate with VEGF expression but not with PTEN expression in squamous cell carcinoma of the larynx. Diagn Pathol. 2010;5:35. | ||
Chen YF, Luo RZ, Li Y, et al. High expression levels of COX-2 and P300 are associated with unfavorable survival in laryngeal squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2013;270(3):1009–1017. | ||
Bayazit YA, Buyukberber S, Sari I, et al. Cyclo-oxygenase 2 expression in laryngeal squamous cell carcinoma and its clinical correlates. ORL J Otorhinolaryngol Relat Spec. 2004;66(2):65–69. | ||
Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–8707. | ||
Chiang Y, Song Y, Wang Z, et al. Aberrant expression of miR-203 and its clinical significance in gastric and colorectal cancers. Gastrointest Surg. 2011;15(1):63–70. | ||
Tian L, Li M, Ge J, et al. MiR-203 is downregulated in laryngeal squamous cell carcinoma and can suppress proliferation and induce apoptosis of tumours. Tumour Biol. 2014;35(6):5953–5963. | ||
Hailer A, Grunewald TG, Orth M, et al. Loss of tumor suppressor mir-203 mediates overexpression of LIM and SH3 Protein 1 (LASP1) in high-risk prostate cancer thereby increasing cell proliferation and migration. Oncotarget. 2014;5(12):2918–4153. | ||
Shibata M, Kodani I, Osaki M, et al. Cyclo-oxygenase-1 and -2 expression in human oral mucosa, dysplasias and squamous cell carcinomas and their pathological significance. Oral Oncol. 2005;41(3):304–312. | ||
Mohan S, Epstein JB. Carcinogenesis and cyclooxygenase: the potential role of COX-2 inhibition in upper aerodigestive tract cancer. Oral Oncol. 2003;39(6):537–546. | ||
Gallo O, Masini E, Bianchi B, Bruschini L, Paglierani M, Franchi A. Prognostic significance of cyclooxygenase-2 pathway and angiogenesis in head and neck squamous cell carcinoma. Hum Pathol. 2002;33(7):708–714. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.