Back to Journals » Cancer Management and Research » Volume 10
MicroRNA-200b expression level is negatively associated with pathological grading in human gliomas
Authors Kong X, Gong S, Yan T, Yang Y
Received 14 April 2018
Accepted for publication 18 June 2018
Published 24 August 2018 Volume 2018:10 Pages 2825—2834
DOI https://doi.org/10.2147/CMAR.S171137
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Antonella D'Anneo
Xiangyi Kong,1,2,* Shun Gong,3–5,* Tao Yan,6,* Yi Yang1
1Department of Neurosurgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences, Beijing 100730, People’s Republic of China; 2Department of Breast Surgical Oncology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, People’s Republic of China; 3Department of Neurosurgery, The General Hospital of Shenyang Military, Army Institute of Neurology, Shenyang 110016, People’s Republic of China; 4Department of Neurosurgery, Shanghai Institute of Neurosurgery, PLA Institute of Neurosurgery, Changzheng Hospital, Second Military Medical University, Shanghai 200003, People’s Republic of China; 5Department of Radiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA; 6Department of Anesthesiology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, People’s Republic of China
*These authors contributed equally to this work
Aim: To elucidate the clinical implication of microRNA (miRNA)-200b in the pathological grading of gliomas.
Methods: We searched the Chinese National Knowledge Infrastructure, Web of Knowledge, Embase, and PubMed databases. Related articles were assessed, and ORs with 95% CIs were calculated to examine the relationship between miRNA-200b expression levels and the World Health Organization (WHO) glioma grade, patients’ sex and age, tumor size, and extent of surgical resection. Heterogeneity, publication bias, and stability of the pooled results of the included studies were also analyzed. MiR-200b expression in 87 human glioma tissues (50 high grade and 37 low grade) and matched 41 non-neoplastic brain tissues was measured by real-time quantitative RT-PCR assay.
Results: Five eligible studies involving 630 patients were included in the present meta-analysis. The miRNA-200b expression in glioma tissues was negatively associated with the WHO glioma grade (OR, 0.070; 95% CI, 0.007–0.678; P=0.022). No significant correlations were found between miRNA-200b and sex (P=0.858), age (P=0.776), tumor size (P=0.134), or extent of resection (P=0.778). In our own test, compared with non-neoplastic brain tissues, the expression level of miR-200b was significantly decreased in glioma tissues (tumor vs normal: 4.29±1.90 vs 10.45±2.34, P<0.001). In addition, we found that the glioma tissues from high-grade tumors (grade III and IV) had much lower miR-200b expression than glioma tissues from low grade tumors (grade I and II).
Conclusion: Our results suggest that the miRNA-200 expression level may be negatively associated with the WHO glioma grade (malignancy). MiRNA-200 might serve as a prognostic and diagnostic biomarker or a helpful therapeutic target.
Keywords: microRNA-200b, down-regulation, glioma, grade, meta-analysis
Introduction
Gliomas can be classified into four pathological grades based on the 2007 World Health Organization (WHO) classification. Low-grade glioma refers to WHO grade I and II gliomas, the prognosis of which is better than that of high-grade gliomas (WHO grade III and IV).1 Although the treatment of choice for gliomas has been extensively researched, current therapies generally remain incapable of significantly improving the prognosis and survival time of patients with gliomas. The main obstacle to treatment efficacy is our insufficient knowledge about the biological molecular mechanisms of malignant gliomas.2
In recent years, microRNA (miRNA) has been recognized to play important roles in the post-transcription regulation of gene expressions. The miRNA-200 family is transcribed from two chromosomal locations: chromosome 1p36, which contains miRNA-200b, miRNA-200a, and miRNA-429, and chromosome 12p13, which contains miRNA-200c and miRNA-141.3 Decreased expression of miRNA-200 has been proven in gastric, lung, endometrial, ovarian, and breast cancers.4–8 MiRNA-200b is recognized as a crucial regulator of chemosensitivity, epithelial–mesenchymal transition, and the cell cycle.9 Several recent studies have demonstrated lower expression of miRNA-200b in malignant glioma specimens and cell lines.10–14 Peng et al reported that over-expression of miR-200b inhibited the colony formation and proliferation of glioma cells.15 Oncogene-encoding CREB1, a significant transcription factor involved in the metastasis, survival, and proliferation of cancer cells, has been confirmed as a direct target-gene of miR-200b. They also demonstrated that CREB1 was present at high levels in glioma tissues and was inversely related to miR-200b expressions. CREB1’s ectopic expressions suppressed the growth-suppressive phenotypes of glioma cells due to miR-200b. Nevertheless, the detailed relationship between miRNA-200b and the glioma WHO grade remains unclear and under discussion, partly because of the small sample sizes in previous studies and the potential heterogeneity among these studies. We performed the present meta-analysis to elucidate the more exact clinical implications of miRNA-200b in the pathological grading of gliomas. We also collected glioma samples and normal brain tissue samples to test the levels of miRNA-200b.
Methods
Search strategy, study selection, and data extraction
The ethics committee of Changzheng Hospital, Second Military Medical University reviewed and approved our research.
Two authors independently conducted computer searches of PubMed, Web of Knowledge, Embase, and Chinese National Knowledge Infrastructure (CNKI) to collect articles describing case–control studies related to the association between miRNA-200b and the risk of glioma. Search terms involved “microRNA-200”, “glioma [MeSH]”, “expression”, and “grade”. The relevant reference articles in the identified studies were reviewed to identify other potentially eligible publications. The inclusion and exclusion criteria are listed in Table 1.
  | Table 1 Inclusion criteria for study selection in this meta-analysis Abbreviations: WHO, World Health Organization; miRNA, microRNA; PCR, polymerase chain reaction; OR, odds ratio. |
The two investigators independently extracted necessary data from all eligible publications according to the inclusion criteria listed in Table 1. The following information was abstracted from each of the included articles: names of the authors, publication year, study population, journal name, mean age, patient numbers, histopathologic findings, test methods, WHO grades, and miRNA-200b level in surgical specimens. Disagreements between the two reviewers were resolved by a third investigator.
Data synthesis and analysis
We conducted the meta-analysis according to the PRISMA checklist and followed the guideline. Differences were expressed as odds ratios (ORs) with 95% CIs. L’Abbé plots, Cochran Q-tests, and I2 tests (variations of OR attributable to heterogeneity) (Table 2) were conducted to estimate the heterogeneity among the included studies.16,17 Subgroup analyses were not performed because of the limited number of included studies. If no significant heterogeneity among the studies was detected, a fixed-effects model was adopted. Otherwise, we used the random-effects model described by Laird and DerSimonian. Sensitivity analyses (Table 2) were conducted to assess the stability of the pooled results.17 Publication bias was evaluated using contour-enhanced funnel plots18 (Table 2). Because only five studies were included, we did not perform a meta-regression analysis.
  | Table 2 The statistical methods used in this meta-analysis and their explanation |
A P-value of <0.05 was considered statistically significant. Data analyses were conducted using STATA 12.0 (StataCorp LP, College Station, TX, USA) and Review Manager 5.3 software (Nordic Cochrane Center, The Cochrane Collaboration, Copenhagen, Denmark).
Further investigation and confirmation
Patients and tissue samples
Written informed consent was obtained from all the patients. All specimens were handled and made anonymous according to the ethical and legal standards. A total of 87 human gliomas (50 high-grade [WHO grade III and IV] and 37 low-grade [WHO grade I and II]) and 41 age- and sex-matched non-neoplastic brain tissues obtained from patients with primary gliomas were collected between 2015 and 2017. The patients with glioma included 55 females and 73 male patients ranging in age from 27 to 76 years (mean, 51 years). All patients underwent MRI a few days before and within 72 hours after surgery. The extent of tumor resection was determined using the postoperative MRI scans. Surgical resection was defined as macroscopic total resection, partial resection or biopsy. According to WHO criteria, 37 of the 87 gliomas were classified as low-grade (WHO grade I and II), and 50 were classified as high-grade gliomas (WHO grade III and IV). None of the patients had received chemotherapy or radiotherapy prior to surgery. Each non-neoplastic brain tissue sample was acquired a minimum of 3.5 cm from the tumor tissues. Resected tissue samples were immediately cut and snap-frozen in liquid nitrogen before being stored at –80°C until RNA extraction.
Real-time quantitative RT-PCR for miRNA
MiR-200b expression in 266 pairs of human gliomas and matched non-neoplastic brain tissues was measured by real-time quantitative RT-PCR assay. Similar to our previous studies, total RNA was extracted from tissue samples using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Each sample was examined in triplicate. Mean normalized gene expression1 SD was calculated from independent experiments. The PCR primers were designed as follows: miR-200b sense, 5′-GCG GCT AAT ACT GCC TGG TAA-3′, and reverse, 5′-GTG CAG GGT CCG AGG T-3′; U6 sense, 5′-CGC TTC GGC AGC ACA TAT ACT A-3′, and reverse, 5′-CGC TTC ACG AAT TTG CGT GTC A-3′.
Statistical analysis
All computations were carried out using SPSS version 13.0 for Windows software (SPSS Inc., Chicago, IL, USA). Data were expressed as mean ± SD. The chi-squared test was used to assess miR-200b expression with respect to clinicopathological parameters. Differences were considered statistically significant when P was less than 0.05.
Results
Search results and study characteristics
The literature search was conducted as presented in Figure 1. A total of five articles finally met the inclusion criteria.10–14 All five studies were performed in Asian populations. One was written in Chinese14 and four were written in English.10–13 The baseline characteristics of the studies are shown in Table 3. The miRNA-200b levels were measured using real-time PCR in four studies and in situ hybridization in one study. Standards and cut-off values used to evaluate the miRNA-200b level are listed in Table 4.
  | Figure 1 Literature search and selection of articles. Abbreviation: miRNA, microRNA. |
  | Table 4 Standards or cut-off values used to evaluate the expression level of microRNA (miRNA)-200b in glioma surgical specimens |
Meta-analysis of relationship between miRNA-200b and gliomas
All gliomas were divided into high grade (WHO grade III + IV) and low grade (WHO grade I+II) for the data analyses. In total, 244 patients with low-grade gliomas and 386 patients with high-grade gliomas were analyzed in this study. The points on the L’Abbé plot did not exhibit a linear distribution (Figure 2), suggesting heterogeneity among the included studies (Q=95.83, df=4, I2=95.8%, P<0.001). The heterogeneity was relatively high; therefore, we pooled the data using a random-effects model, which assumes that the data being analyzed are drawn from a hierarchy of different populations whose differences relate to that hierarchy, assisting in controlling for unobserved heterogeneity. As revealed in Figure 3A, the forest plot demonstrated that the expression of miR-200b was much higher in low-grade than high-grade gliomas. The integrated ORs suggested a significant negative correlation between the miRNA-200b expression level and the WHO glioma grade (OR, 0.070; 95% CI, 0.007–0.678; P=0.022). We also tested the correlation between the miRNA-200b level and sex, age, tumor size, and extent of resection. Figure 2B and C shows that the points on the L’Abbé plots basically exhibited a linear distribution, providing evidence of homogeneity among the studies and suggesting the rationality of using a fixed-effects model. The five studies involved 335 male and 295 female patients and 293 young and 337 old patients. No significant associations were found between the miRNA-200b level and either sex (OR, 1.029; 95% CI, 0.749–1.415; P=0.858) or age (OR, 1.048; 95% CI, 0.760–1.445; P=0.776) (Figure 3B and C). Only two studies focused on tumor size and the extent of surgical resection, and no significant differences were detected (tumor size: OR, 0.715; 95% CI, 0.462–1.108; P=0.134 and extent of resection: OR, 1.072; 95% CI, 0.660–1.742; P=0.778) (Figure 3D and E).
Sensitivity analysis and publication bias
Each individual study was deleted one at a time to determine whether the study affected the pooled ORs. Our results showed that the ORs were not affected by omitting the included studies (Figure 2D–F). Contour-enhanced funnel plots were adopted to reflect publication-bias and showed that the literature had missing areas of high statistical significance (right-hand side of the plots), suggesting no publication bias (Figure 2G–I).
Further investigation and confirmation of results
The expression levels of miR-200b in all glioma tissues with WHO grade I–IV and non-neoplastic brain tissues are shown in Figure 4. Compared with non-neoplastic brain tissues, the expression level of miR-200b was significantly decreased in glioma tissues (tumor vs normal: 4.29±1.90 vs 10.45±2.34, P<0.01). Additionally, a significant relationship was observed between miR-200b expression and the histopathologic grade. The glioma tissues from high-grade tumors (grade III and IV, 3.13±0.89) had much lower miR-200b expression than glioma tissues from low-grade tumors (grade I and II, 5.87±1.77) (P<0.01).
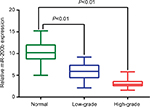  | Figure 4 MiR-200b expressions in glioma tissues and non-neoplastic brain tissues were respectively detected by real-time quantitative RT-PCR assay. Abbreviation: miRNA, microRNA. |
Discussion
In the present study, we identified five studies that investigated the effects of miRNA-200b level on the WHO glioma grade. We found that miRNA-200b level was negatively associated with WHO glioma grade. However, no correlations were found between the miRNA-200b level and sex, age, tumor size, or extent of surgical resection. To our knowledge, this is the first meta-analysis to systematically estimate the relationship between miRNA-200b and the WHO glioma grade.
Some studies have shown that the miRNA-200 family plays a tumor-suppressive role in a wide range of human cancers, including endometrial carcinoma, breast cancer, colorectal cancer, pancreatic cancer, and gastric cancer.4–8 Interestingly, Kahlert et al19 revealed that the Zeb1–miR-200 feedback loop could interconnect the processes of invasion, chemoresistance, and tumorigenesis in glioblastoma multiforme through the downstream effectors ROBO1, c-MYB, and MGMT. Recent studies also have suggested that miRNA-200b is frequently down-regulated in glioma, and the decreased miRNA-200b expression is an independent unfavorable prognostic factor for patients with glioma. Additionally, miRNA-200b has been reported to play a suppressive role in the regulation of glioma cell growth by targeting CREB1. Liu et al10 suggested that miRNA-200b has suppressive effects on the proliferation, migration, invasion, and epithelial–mesenchymal transition of glioma cells by targeting Zeb2, a DNA-binding transcriptional repressor that plays a role in the TGF-β signaling pathways that are essential during early cancer development. In a luciferase reporter assay, Li et al20 also found that miRNA-200b inhibits the growth and metastasis of glioma cells by targeting ZEB2. Nevertheless, few studies have systematically described the precise clinical implications of miRNA-200b in glioma development and its precise roles in WHO grading, and the sample size in each study was not large enough. As such, whether miRNA-200b can be used as a dependable biomarker for WHO glioma grading remains unclear. We therefore combined the search results of PubMed, Web of Knowledge, Embase, and CNKI databases to systematically analyze the clinical implications of miRNA-200b.
We explored the correlation between miRNA-200b expression and the WHO glioma grade in 630 patients from five studies that investigated the pathologies in glioma specimens. We found that miRNA-200b was negatively correlated with higher WHO grades in patients with gliomas. Specifically, the pooled OR and 95% CI showed a significant negative correlation between miRNA-200b and WHO grade (n=5; OR, 0.070; 95% CI, 0.007–0.678; P=0.022). There were no significant correlations between miRNA-200b and sex (P=0.858), age (P=0.776), tumor size (P=0.134), or the extent of resection (P=0.778).
This finding that miR-200b is down-regulated in glioma is consistent with the findings reported by Peng et al.21 They found an inverse correlation between the miR-200b level and CREB1 protein level.21 MiR-200b directly targets the 3’-untranslated region of the CREB1 transcript and elicits specific and robust knockdown of CREB1 protein.21 Hu et al22 investigated the expression and function of LDHA in patients with gliomas. They found that LDHA was up-regulated in glioma specimens and that it promoted proliferation, invasion, and glycolysis in glioma cells. They also performed luciferase reporter assays and confirmed that LDHA was a direct target of miR-200b, the expression of which was inversely correlated with LDHA expression.22 The results of the present meta-analysis are also consistent with the findings of our own study. In this study, we obtained frozen tissue samples of 87 glioma tissues and 41 normal brain tissues. The tumor samples were diagnosed by two pathologists who were blinded to the patient data. We adopted quantitative real-time PCR to measure the expression of miR-200b in glioma tissues and normal brain tissues (P<0.01). The results showed that the expression of miR-200b was significantly lower in gliomas than in normal tissues. Additionally, the miR-200b levels in grade I and II gliomas were much higher than those in grade III and IV gliomas (P<0.01). Our results further confirmed this association.
Limitations
Our meta-analysis should be interpreted within the context of its limitations. First, all included studies were performed in Asian populations. Patients of other races should be considered in future studies. Second, whether miRNA-200b represents an independent prognostic indicator beyond its ability to predict the WHO glioma grade remains unclear. A subgroup analysis of established glioma biomarkers, such as isocitrate dehydrogenase mutation, would be clinically meaningful. Third, the expression of other genes or non-coding RNAs that may interact with miRNA-200b should be considered. Fourth, it would be interesting, where possible from meta-analysis data, to verify whether miR-200b is a prognostic factor in terms of overall survival or disease-free survival of patients. An alternative strategy could be the use of data from the The Cancer Genome Atlas (TCGA), which will be our next work in future. Fifth, as is known, large quantities of miRNAs have been reported to be involved in gliomagenesis. More lines of evidence are needed to support our conclusions. And also, the signal-transducing mechanisms explaining roles of miR-200b in gliomagenesis are to be evaluated and verified in our next work. Finally, the cut-off points for miRNA-200b expression differed among the studies, which might have affected our results. Notably, the miR-200 family contains miR-200a, miR-200b, miR-200c, miR-141, and miR-429. Although all miR-200 miRNAs have been reported to be involved in cancer development and metastasis, reports on the influence of members other than miR-200b on glioma are very limited. Peng et al15 reported that miRNA-141 could inhibit glioma cell growth and metastasis by targeting TGF-β2. Sun et al23 recently investigated the expression level, clinicopathological significance, and prognostic significance of miR-429 in gliomas and found an increased miR-429 level (with ascending pathological grade) in glioma tissues compared with non-neoplastic brain tissues (P<0.01). Serna et al24 showed that miRNA-200c and E-cadherin expression are down-regulated while ZEB1 is up-regulated in glioblastomas with a high level of epidermal growth factor receptor (EGFR) amplification. Therefore, more studies are necessary to determine whether the significance of our results is specific to miRNA-200b.
Acknowledgments
This study was supported by grants from Beijing Municipal Science and Technology Project (No. Z151100003915109), Capital Public Health Education, Beijing Science and Technology Program (No. Z171100000417028), and Chinese Academy of Medical Sciences Initiative for Innovative Medicine (No. 2017-I2M-3-020). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
XK and SG searched the literature and analyzed the data; TY also analyzed the data and drafted some figures; YY put forward the idea. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Brat DJ, Scheithauer BW, Fuller GN, Tihan T. Newly codified glial neoplasms of the 2007 WHO Classification of Tumours of the Central Nervous System: angiocentric glioma, pilomyxoid astrocytoma and pituicytoma. Brain Pathol. 2007;17(3):319–324. | ||
Killock D. CNS cancer: molecular classification of glioma. Nat Rev Clin Oncol. 2015;12(9):502. | ||
Paterson EL, Kazenwadel J, Bert AG, Khew-Goodall Y, Ruszkiewicz A, Goodall GJ. Down-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progression. Neoplasia. 2013;15(2):180–191. | ||
Cristóbal I, Rincón R, Manso R, et al. Deregulation of miR-200b, miR-200c and miR-429 indicates its potential relevant role in patients with colorectal cancer liver metastasis. J Surg Oncol. 2014;110(4):484–485. | ||
Kurashige J, Mima K, Sawada G, et al. Epigenetic modulation and repression of miR-200b by cancer-associated fibroblasts contribute to cancer invasion and peritoneal dissemination in gastric cancer. Carcinogenesis. 2015;36(1):133–141. | ||
Li A, Omura N, Hong SM, et al. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70(13):5226–5237. | ||
Pan Y, Liang H, Chen W, et al. microRNA-200b and microRNA-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs. RNA Biol. 2015;12(3):276–289. | ||
Yoneyama K, Ishibashi O, Kawase R, Kurose K, Takeshita T. miR-200a, miR-200b and miR-429 are onco-miRs that target the PTEN gene in endometrioid endometrial carcinoma. Anticancer Res. 2015;35(3):1401–1410. | ||
Liu W, Qi L, Lv H, et al. MiRNA-141 and miRNA-200b are closely related to invasive ability and considered as decision-making biomarkers for the extent of PLND during cystectomy. BMC Cancer. 2015;15:92. | ||
Liu Q, Tang H, Liu X, et al. miR-200b as a prognostic factor targets multiple members of RAB family in glioma. Med Oncol. 2014;31(3):859. | ||
Wang B, Li M, Wu Z, et al. Associations between SOX2 and miR-200b expression with the clinicopathological characteristics and prognosis of patients with glioma. Exp Ther Med. 2015;10(1):88–96. | ||
Li J, Yuan J, Yuan X, et al. MicroRNA-200b inhibits the growth and metastasis of glioma cells via targeting ZEB2. Int J Oncol. 2016;48(2):541–550. | ||
Men D, Liang Y, Chen L. Decreased expression of microRNA-200b is an independent unfavorable prognostic factor for glioma patients. Cancer Epidemiol. 2014;38(2):152–156. | ||
Shen D, Liu T, Lin Q, et al. MGMT promoter methylation correlates with an overall survival benefit in Chinese high-grade glioblastoma patients treated with radiotherapy and alkylating agent-based chemotherapy: a single-institution study. PLoS One. 2014;9(9):e107558. | ||
Peng T, Zhang S, Li W, Fu S, Luan Y, Zuo L. MicroRNA-141 inhibits glioma cells growth and metastasis by targeting TGF-β2. Am J Transl Res. 2016;8(8):3513–3521. | ||
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. | ||
Dickersin K, Berlin JA, Meta-analysis: state-of-the-science. Epidemiol Rev. 1992;14:154–176. | ||
Stuck AE, Rubenstein LZ, Wieland D. Bias in meta-analysis detected by a simple, graphical test. Asymmetry detected in funnel plot was probably due to true heterogeneity. BMJ. 1998;316(7129):469; author reply 470-1. | ||
Kahlert UD, Suwala AK, Raabe EH, et al. ZEB1 Promotes Invasion in Human Fetal Neural Stem Cells and Hypoxic Glioma Neurospheres. Brain Pathol. 2015;25(6):724–732. | ||
Li J, Yuan J, Yuan X, et al. MicroRNA-200b inhibits the growth and metastasis of glioma cells via targeting ZEB2. Int J Oncol. 2016;48(2):541–550. | ||
Peng B, Hu S, Jun Q, et al. MicroRNA-200b targets CREB1 and suppresses cell growth in human malignant glioma. Mol Cell Biochem. 2013;379(1–2):51–58. | ||
Hu S, Jiang Q, Luo D, et al. miR-200b is a key regulator of tumor progression and metabolism targeting lactate dehydrogenase A in human malignant glioma. Oncotarget. 2016;7(30):48423–48431. | ||
Sun X, Li Z, Chen Y. The Potential Prognostic Value of MicroRNA-429 for Human Gliomas. Ann Clin Lab Sci. 2016;46(1):44–48. | ||
Serna E, Lopez-Gines C, Monleon D, et al. Correlation between EGFR amplification and the expression of microRNA-200c in primary glioblastoma multiforme. PLoS One. 2014;9(7):e102927. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



