Back to Journals » Cancer Management and Research » Volume 11
MicroRNA-17 promotes cell proliferation and migration in human colorectal cancer by downregulating SIK1
Authors Huang C, Liu J, Xu L , Hu W , Wang J , Wang M , Yao X
Received 7 November 2018
Accepted for publication 5 March 2019
Published 24 April 2019 Volume 2019:11 Pages 3521—3534
DOI https://doi.org/10.2147/CMAR.S191087
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Kenan Onel
Chengzhi Huang,1,2 Jianhua Liu,3 Lishu Xu,4 Weixian Hu,1,5 Junjiang Wang,1,5 Muqing Wang,1,6 Xueqing Yao1–2,5–6
1Department of General Surgery, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, Guangdong, People’s Republic of China; 2Medical College, Shantou University, Shantou, Guangdong, People’s Republic of China; 3Department of Gastroenterology Oncology, Cancer Center, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, Guangdong, People’s Republic of China; 4Department of Gastroenterology, Guangdong Provincial Geriatrics Institute, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, Guangdong, People’s Republic of China; 5The Second School of Clinical Medicine, Southern Medical University, Guangzhou, People’s Republic of China; 6School of Medicine, South China University of Technology, Guangzhou, Guangdong, People’s Republic of China
Purpose: There is mounting evidence to indicate that microRNA-17 (miR-17) is expressed and functionally involved in human cancers. However, the molecular mechanism underlying the role of miR-17 in colorectal cancer (CRC) remains largely unclear. This study aims to reveal the biological function of miR-17 in colorectal cancer.
Materials and methods: The expression of miR-17 in CRC cells and tissues was examined using qRT-PCR. Cell proliferation and migration assays were performed after transfection with an miR-17 mimic and inhibitors. The potential gene targets of miR-17 were predicted by bioinformatics analysis and further validated by PCR, Western blot and dual luciferase reporter assays.
Results: The expression of miR-17 was significantly upregulated in CRC cell lines and tissues and may imply poor prognosis. miR-17 upregulation promoted cell invasion and migration in CRC cell lines in vitro, while downregulation of miR-17 inhibited tumor progression. SIK1 was identified as a potential direct target of miR-17 by dual luciferase reporter assay, and its downregulation in CRC may suggest poor prognosis.
Conclusions: Our study indicated that upregulated miR-17 may promote the progression of CRC and may exert its function as a tumor suppressor miRNA by targeting SIK1.
Keywords: SIK1, colorectal cancer, prognosis, tumor biomarker
Introduction
Colorectal cancer (CRC) is one of the most common malignant diseases in the world. In recent years, significant improvements have been made in several CRC treatment options, including surgical resection techniques, chemotherapy and immunotherapy therapy. However, the 5-year overall survival rate of CRC still remains low, due to the tumor invasion and metastasis after treatment.1-4
Numerous studies have focused on the identification of clinical factors that may predict treatment outcomes for patients with CRC. It has been demonstrated that histopathological factors, such as tumor size, the degree of cancer differentiation, as well as lymph vascular invasion and lymph node involvement may act as prognostic factors for CRC patients.5 However, increasing evidence suggests that CRC patients with the same tumor stage may have significantly different prognoses due to tumor heterogeneity.6 Therefore, it is necessary to identify novel tumor biomarkers to refine risk stratification and survival prognosis of CRC patients.
MicroRNAs (miRNAs), approximately 22 nucleotides in length, have been shown to post-transcriptionally regulate gene expression by binding to the seed region and sequence in the 3ʹ-untranslated region (UTR) of its target mRNA. MicroRNAs may also lead to translational repression or degradation of target mRNAs. Several studies have shown that aberrant expression of miRNAs, such as miR-21, miR-155, miR-223, and miR-519d, may be involved in the processes of cell proliferation, metastasis, and chemoresistance in human cancers.7–10 In our pervious study, we found that increased expression of miR-17 played an unfavorable role in overall survival (OS) in human malignancy.11 Studies have shown that miR-17 may regulate cervical cancer progression by regulating p53-induced nuclear protein 1.12 Although several studies have suggested that aberrant expression of miR-17 may be involved in the progression and metastasis of CRC, its functional role in CRC has not been established.
Salt-inducible kinase 1 (SIK1) encodes a serine kinase of the AMP-activated protein kinase family, which plays critical roles in cell growth and metabolism.13,14 A previous study has demonstrated that SIK1-knockdown cells can escape anoikis in metastatic cancer tissues.15 Additionally, SIK1 may lead to p53-dependent anoikis and subsequent suppression of tumor cell metastasis.16 Other related studies have shown that reduced expression of SIK1 may also be associated with tumor progression, metastasis, and poor clinical outcome in ovarian cancer, breast cancer, pancreatic cancer, and hepatocellular carcinoma.15–18 Therefore, SIK1 is classified as a tumor suppressor gene in many cancers. However, the status of SIK1 expression and its clinical significance in CRC remains unclear.
In the present study, we first examined SIK1 expression in tumor tissues from 101 CRC patients by immunohistochemistry (IHC) analysis, Western blot, and qRT-PCR, as well as its association with clinicopathological features of CRC. Secondly, the biological function of miR-17 in CRC was investigated in vitro. Additionally, our analysis also confirmed that SIK1 is a direct target gene of miR-17. Finally, we hypothesize that SIK1 mRNA may be a direct target of miR-17 and that SIK1 protein may be a prognostic biomarker and potential therapeutic target for CRC.
Materials and methods
Patients, follow-up and tissue samples
A total of 101 patients with CRC who had underwent surgical colorectal resection in the Department of General Surgery, Guangdong Provincial People’s Hospital (Guangdong Academy of Medical Sciences), between June 2012 and January 2013 were enrolled in this study.
The patients enrolled in this study met the following requirements: a pathology-confirmed diagnosis of colorectal cancer adenocarcinoma; patients with stage I-IV cancer who did not receive any radiotherapy or chemotherapy before surgical resection. TNM staging was based on the guidelines of the 7th version of the AJCC colon and rectum cancer staging manual. All enrolled patients were followed up by out-patient department evaluation or telephone interview until death up to July 2017. The patients ranged from 22 to 92 years old, with a mean age of 60.75±1.42 years old. IHC analysis was conducted to evaluate the altered expression level of SIK1 protein in paired tissues from 101 enrolled patients. qRT-PCR was conducted to evaluate the expression of miR-17 in tumor and non-tumor tissues.
Immunohistochemistry analysis
Paraffin-embedded sections (5 µm) of cancerous tissue were heated in a 60 °C baker for 30 min. The sections were then deparaffinized with xylene and rehydrated, followed by treatment with 3% hydrogen peroxide in methanol. Then, non-specific binding was blocked using 1% bovine serum albumin. Sections were then incubated with a SIK1 antibody (1:500, ab64428, Abcam, Cambridge, MA, USA) overnight at 4 °C. Incubation with PBS was used as a negative control. After washing, sections were then incubated with a biotinylated secondary antibody at room temperature. DAB was used to visualize positive staining. Slides were then dehydrated and sealed with coverslips. Two independent investigators without prior knowledge of the patients were responsible for evaluating the immunohistochemistry results. Using a light microscope, cells in 5 randomly selected fields were quantified on 3 slides for each specimen. The expression of SIK1 was determined based on the staining intensity as well as the percentage of positive cells. The staining data was processed using Image J software.19 The staining scores were defined as follows: 1 (<10% positive cells), 2 (10–50% positive cells), 3 (50–75% positive cells), and 4 (>75% positive cells). The staining intensity was determined by the following standards: 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, brown), and 3 (strong staining, brown). The final score of SIK1 expression was the combination of the staining intensity and percentage of positive cells, for an aggregate score of 0 for negative, + for 1–3 points, ++ for 4–6 points, and +++ for 7–9 points. Low expression was defined as 0 or +, while ++ and +++ were considered high expression. Cut-off values for SIK1 were chosen based on the measure of heterogeneity by using the log-rank test with consideration of overall survival time.
Cell culture and transfection
Human colorectal cancer cell lines HCT116, SW480, and HT29 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Colon epithelial cells were purchased from the iBS Fudan Cell Bank (Shanghai, China). All cell lines were cultured in RPMI-1640 medium (Hyclone, Biowest) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) in a humidified 37 °C incubator with 5% CO2.
MicroRNA 17 (5ʹ- CAAAGUGCUUACAGUGCAG-GUAG-3ʹ) was synthesized by GenePharma (Shanghai, China). The miR-17 mimic (5ʹ-CAAAGUGCUUACAG-UGCAGGUAG-3ʹ), negative-control (NC) mimic (5ʹ-UC-ACAACCUCCUAGAAAGAGUAGA-3ʹ), miR-17 inhibitor (5ʹ-CUACCUCGCACUGUAAGCACUUUG-3ʹ), and NC inhibitor (5ʹ-UUGUACUACAAAAGUACUG-3ʹ) were purchased from GenePharma (Shanghai, China). For transfections, SW480 and HCT116 cells were plated in a 6-well plate (3.0×105 per well) and the cells were cultured in complete serum medium until the confluence reached 40–50%. The cells were then transfected with 50 nM miRNA using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions. After transfection (48 hrs), the cells were harvested for further investigations.
Quantitative real-time PCR
For the analysis of miR-17, total miRNA from frozen tissues was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The extracted mRNA was then used to synthesize cDNA using a Takara miRNA First-Strand Synthesis Kit. U6 was used for miR-17 normalization, and the comparative Ct method was used to calculate the relative abundance of miR-17. Total RNA was extracted from surgical specimens or cultured cells using TRIzol (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized using a Takara mRNA First-Strand Synthesis Kit. SIK1 expression level was quantified using SYBR Green PCR master mix (Roche, USA) on the Applied Biosystems 7500 (Life Technologies). All tests were performed in triplicate. The primer sequences used are as follows:
miR-17 Forward (F): 5ʹ-ACACTCCAGCTGGGCAA-AGTGCTTACAGTGC-3ʹ
Reverse(R): 5ʹ-TGGTGTCGTGGAGTCG-3ʹ
SIK1 mRNA Forward (F): 5ʹ->TTCCGGCAGCAGC-TAGCGGCAAACGCGAGGACCAAG-3ʹ
Reverse (R): 5ʹ-CTAAGGAAAAACGCGGCGACCG-CGGGGTTCCTGGGACTG-3ʹ.
U6 Forward (F): 5ʹ-CTCGCTTCGGCAGCACA-3ʹ
Reverse (R): 5ʹ-AACGCTTCACGAATTTGCGT −3ʹ.
GAPDH Forward (F): 5ʹ-CTACAATGAGCTGCGTG-TGG-3ʹ
Reverse (R): 5ʹ-AAGGAAGGCTGGAAGAGTGC-3ʹ.
Protein extraction and Western blot
Colorectal cancer tissue samples and cells were harvested and then lysed using RIPA buffer (CWbiotech, Beijing, China) supplemented with complete protease inhibitor cocktail (CWbiotech, Beijing, China). The extracted protein was heated with SDS-PAGE buffer (CWbiotech, Beijing, China). Equal amounts of protein were separated on SDS polyacrylamide gels and transferred to PVDF membranes (Immobilon P, Millipore). The membranes were then blocked in fat-free milk in TBS with 0.1% Tween-20 (Sangon Biotech, Shanghai, China) for 60 min at room temperature. The membranes were then incubated with an anti-SIK1 antibody (1:1000, ab64428, Abcam, Cambridge, MA, USA) overnight at 4 °C. The HRP-conjugated anti-rabbit secondary antibody (1:5000, ab6721, Abcam, Cambridge, MA, USA) was administered for 60 min at room temperature. Each assay was repeated three times. The membranes were scanned using Syngene Imaging System (Frederick, MD, USA). The expression of SIK1 protein was measured by ImageJ based on the grey density ratio.19
Cell counting kit-8 (CCK-8) assay
The proliferation of cells transfected with miR-17 mimic and miR-17 inhibitors were evaluated using the CCK-8 detection kit (Beyotime Biotechnology, Shanghai, China) according to the manufacturer’s instructions. The cells were incubated in a 96-well microplate at a density of 5×104 cells/ml, and 24 hrs later, cells were treated with resveratrol. The CCK-8 solution (20 ml/well) was then added to the cells, which were then incubated at 37 °C for 2 hrs. The absorbance of the cells at 450 nm was then measured.
Cell apoptosis assay
Cell apoptosis was assessed using the Cell Apoptosis Kit (Life technologies). Cultured cells were trypsinized and then incubated with Annexin V-FITC and PI solution for 15 mins. The cell samples were then analyzed with a CellStream Flow Cytometer (Millipore).
Transwell assay
In our study, the invasion of cancer cells was analyzed using the Transwell assay. Approximately 5×105 cells were plated in the upper chambers of 8 µm transwell inserts (BD Biosciences, USA) in serum-free media. Then the lower chambers were filled with normal growth medium. The cells were incubated for 24 hrs allowing for invasion. Cells that migrated to the lower chambers were washed with PBS, mixed with 20% methanol and stained with 0.1% crystal violet. This assay was repeated three times, and the number of stained cells in five randomly chosen fields was determined under a microscope.
Bioinformatics analysis
The comparison of miR-17 expression in CRC tissue and normal tissue was performed using the Oncomine database (
Luciferase reporter assay
The wild-type (WT) 3ʹ UTR sequence of SIK1 was amplified using AmpliTaq Gold DNA Polymerase (Applied Biosystems). The purified 3ʹUTR was then inserted into psiCHECK2 vector (Promega) according to the manufacturer’s instructions. For mutant constructs, the miRNA seed region of the WT 3ʹUTR was mutated using AccuPrime Pfx SuperMix (Invitrogen). Sequencing was used to verify the sequence of the WT and mutant. The co-transfection of SW480 cells with WT or mutant constructs or negative controls was performed using Lipofectamine 2000 (Invitrogen). After 24 hrs, the Dual-Luciferase Report Assay (Promega) was performed. The renilla luciferase activity was normalized to the corresponding firefly luciferase activity.
Statistical analysis
The data shown are mean±standard deviation (SD) based on 3 independent experiments. The student’s independent t-test and one-way ANOVA were used to compare two groups and multiple groups, respectively. The association of SIK1 and clinicopathological features was assessed using χ2 test. The cumulative overall survival was estimated using Kaplan-Meier analysis.28 The survival curves were analyzed by the log-rank test. Overall survival (OS) was determined as the time from tumor resection to the time of patient death. The influence of variables on survival was analyzed by multivariate analysis using COX proportional hazard model. P-value<0.05 was considered statistically significant. All the included data were analyzed using SPSS version 17.0 (SPSS, Chicago, IL, USA).
Ethics approval and patient consent
All enrolled patients provided signed informed consent prior to sample collection. The study protocol conformed to the ethical guidelines outlined in the Declaration of Helsinki and was approved by the research ethics committee of Guangdong Provincial People’s Hospital (Guangdong Academy of Medical Sciences) (approval No.2010011H).
Results
miR-17 expression is upregulated in CRC tissue and may imply poor prognosis
In our previous study, we found that miR-17 may be an oncogene in human malignancies, including colorectal cancer. The elevated expression of miR-17 in tumor tissue may imply poor clinical outcome.11 We further used the online Oncomine database to examine the differential expression of miR-17 in tumor and adjacent non-tumor tissues. Data from a total of 5 independent microarray databases were included in our study. We found that miR-17 is overexpressed in CRC tissues in comparison to normal tissues in the TCGA Colorectal, Gaedcke Colorectal, Kaiser Colon, Skrzypczak Colorectal, and Hong Colorectal databases (Figure 1A). Additionally, qRT-PCR analysis revealed that CRC specimen contained significantly higher levels of miR-17 than normal non-tumor tissues (Figure 1B, P<0.001). In order to further explore the clinical significance of miR-17, 101 patients were followed for 5 years. The median value of all 101 cases was chosen as the cut-off point for delineating miR-17 high (n=51) or low (n=50) expression. In Figure 1C, the Kaplan-Meier curves revealed that CRC patients with higher expression of miR-17 had poor OS (HR=2.336, 95%CI=1.107–4.929, P=0.0206).
miR-17 overexpression promotes migration and invasion of CRC cells
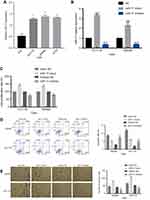
We investigated the expression level of miR-17 in three CRC cell lines in order to further examine its association with CRC. Based on PCR results, the expression of miR-17 was significantly higher in CRC cells than in normal colon cells, FHC (Figure 2A, P<0.05). With relatively higher expression of miR-17, SW480 and HCT116 were chosen for further investigation. SW480 and HCT116 were transfected with miR-17 mimic and miR-17 inhibitor. Quantitative real-time PCR analysis was conducted to confirm that the expression of miR-17 was significantly upregulated in the miR-17 mimic-transfected cells, while the miR-17 inhibitor-transfected cells showed lower expression of miR-17 (Figure 2B, P<0.001). The 24 hr CCK-8 assay was conducted in order to demonstrate that the overexpression of miR-17 promoted the viability of SW480 and HCT116 cells, and the upregulation of miR-17 increased the proliferation of cancer cells. In contrast, cell proliferation was significantly decreased in cells transfected with miR-17 inhibitors (Figure 2C, P<0.05). The flow cytometry assay also demonstrated that the miR-17 mimic transfection decreased apoptosis of HCT116 and SW480 cells, while decreased expression of miR-17 promoted apoptosis (Figure 2D, all P<0.01). We then studied the functional role of miR-17 in the invasion of CRC cells using the Transwell assay. As shown in Figure 2E, invasion was significantly increased in miR-17 mimic-transfected cells, whereas invasion was significantly decreased in cells with decreased expression of miR-17 (all P<0.05). Collectively, these results suggest that miR-17 may contribute to the development and progression of CRC.
SIK1 is a direct target of miR-17 in CRC
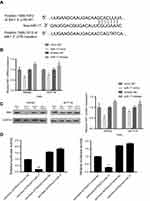
In order to further study the molecular mechanisms of miR-17, online bioinformatics databases, including TargetScan, miRTarbase, and miRWalk were used to predict the potential target genes of miR-17. After screening the databases, 53 potential miR-17 target genes were identified (Figure 3A). After further exploration of TCGA and The Human Protein Atlas, we identified 8 genes (FEM1C, FJX1, PGM2L1, C6orf120, SIK1, FAM117B, PDLIM5, and DNAL1) that may be downregulated in CRC and associated with poor prognosis. This result warranted further consideration of these genes as potential direct targets of miR-17 (Figure 3B). Based on our in silico analyses, we selected SIK1 for further exploration. Using TCGA cohorts and Human Protein Atlas data, we found that downregulation of SIK1 was significantly associated with poor prognosis in CRC (P=0.0031) (Figure 3C). Next, we identified a potential miR-17 binding site in SIK1 (Figure 4A). We then examined SIK1 mRNA and protein expression in both SW480 and HCT116 cells transfected with a miR-17 mimic and miR-17 inhibitor. Quantitative real-time PCR analysis suggested that the overexpression of miR-17 significantly suppressed the expression of SIK1 mRNA, while higher expression of SIK1 mRNA was found in cells transfected with a miR-17 inhibitor (Figure 4B, P<0.01). Similarly, the SIK1 protein expression was found to be repressed in both SW480 and HCT116 cells transfected with miR-17 mimic, while SIK1 was significantly elevated in cells transfected with a miR-17 inhibitor (Figure 4C, all P<0.05). Additionally, a dual luciferase reporter assay was carried out to further examine the relationship between miR-17 and SIK1 in SW480 and HCT116 cells. The SW480 and HCT116 cells were transfected with psiCheck-SIK1-3ʹUTR-WT or psiCheck-SIK1-3ʹUTR-mut along with a miR-17 mimic and NC mimic. The results showed that co-transfection with psiCheck-SIK1-3ʹUTR-WT and psiCheck-SIK1-3ʹUTR-mut led to decreased luciferase activity in both SW480 (Figure 4D, P<0.001) and HCT116 cells (Figure 4E, P<0.001). However, the luciferase activity in SW480 and HCT116 cells transfected with psiCheck-SIK1-3ʹUTR-mut was not affected. These results suggested that SIK1 mRNA may be a direct target of miR-17.
SIK1 expression is downregulated in CRC tissues
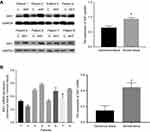
Immunohistochemistry was preformed to evaluate the protein expression of SIK1 in 101 paired colorectal cancer tissues. Positive staining was observed in the cytoplasm. Cancerous tissues exhibited lower expression of SIK1 protein than adjacent normal tissue (P<0.001) (Table 1, Figure 5). To validate the IHC data, qRT-PCR and Western blot assays were performed on randomly chosen sections of patient tissues. The results showed decreased expression of SIK1 protein in the CRC tissue samples (Figure 6A, P<0.05). The qRT-PCR results also showed that SIK1 mRNA was significantly downregulated in tumor tissues in comparison to paired adjacent non-tumor tissues (Figure 6B, P<0.05). To investigate the relevance of SIK1 for predicting clinical outcomes of CRC patients, we then analyzed the correlation between SIK1 expression and the clinicopathological features of CRC patients. As shown in Table 2, immunohistochemistry revealed that the expression of SIK1 was strongly associated with tumor location, differentiation, tumor invasion, lymph node metastasis, and clinical stage (TNM), but did not shown significant association with age, gender, and initial CEA.
 | Table 1 The comparison of SIK1 expression in cancerous tissue and adjacent non-tumor tissue |
 | Table 2 Correlations between SIK1 expression and clinical characteristics of patients with CRC |
SIK1 downregulation is associated with poor overall survival of CRC patients
Patients with lower expression of SIK1 had significantly lower overall survival rates than patients with higher expression of SIK1 (Table 3, P<0.001). The 5-year overall survival rate for all patients was 60.40%. Of the enrolled patients, 40 of them died after the initial surgery. The overall mean follow-up was 2.69 years. Univariate and multivariate analyses of overall survival of the enrolled CRC patients is summarized in Table 3. The Univariate analysis showed that the overall survival after surgical resection was significantly correlated with the following factors: clinical stage (TNM) (P<0.05) and the expression level of SIK1 (P<0.001). Multivariate COX regression analysis revealed that lower SIK1 expression is an independent risk factor for CRC with a hazard ratio (HR) of 0.33 (95% Confidence Interval (CI) 0.16–0.67, P=0.002), as well as clinical stage (TNM), with a hazard ratio of 0.45 (95% CI 0.23–0.89, P=0.021).
 | Table 3 Univariate and multivariate survival analysis of patients with colorectal cancer |
Discussion
The main objective of our study was to explore the role of miR-17 in CRC, as well as identify potential target genes of miR-17. It has been established that miRNAs serve a critical role in the process of oncogenesis by binding to the 3ʹUTR of target genes in multiple malignancies. However, the specific molecular mechanism of miR-17 function in CRC remains unclear. Previous studies have shown that abnormal expression of miR-17 occurs in malignancies, such as pancreatic cancer, lung cancer, breast cancer, and kidney cancer by regulating cell proliferation, tumor growth and epithelial-to-mesenchymal transition.29–33 Previous studies have also suggested that miR-17 may act as an oncogene in CRC.11 Based on our pervious meta-analysis, the upregulation of miR-17 may suggest poor prognosis in cancer patients.11 Additionally, several studies have demonstrated that miR-17 can promote the progression of CRC by directly regulating PTEN and P130, which is consistent with the cell growth and metastatic phenotypes observed in our study.34,35 By using Oncomine and surgical specimens, we demonstrated that the expression of miR-17 was significantly higher in tumor tissues than in adjacent non-tumor tissues. We then validated that the overexpression of miR-17 may indicate poor prognosis of CRC patients. In addition, we confirmed that the overexpression of miR-17 may increase proliferation and invasion of CRC cells, while these functions were impaired in cells transfected with miR-17 inhibitors in vitro. Therefore, the results suggested that miR-17 can promote the progression of CRC cells. In the present study, we also demonstrated that CRC cells transfected with a miR-17 inhibitor were impaired. The results imply that downregulating miR-17 may be a potential therapeutic strategy for CRC patients.
Recently, an increasing number of bioinformatic tools have been developed, which can predict target genes of miRNAs.36 In our study, we screened 3 online databases, including TargetScan, miRTarBase, and miRWalk, and identified 53 common genes predicted by these databases. Additionally, we further explored TCGA and identified 8 potential miR-17 target genes (FEM1C, FJX1, PGM2L1, C6orf120, SIK1, FAM117B, PDLIM5, and DNAL1). Our in silico approach allowed us to identify a potential miR-17 binding site in SIK1. Our data also suggests that downregulation of SIK1 may be associated with poor prognosis. In the present study, we discovered that the expression of miR-17 is inversely associated with the expression of SIK1 in CRC cells. The qRT-PCR assay and Western blot analysis suggested that cells transfected with a miR-17 mimic had lower mRNA or protein expression of SIK1. Additionally, the luciferase reporter assay confirmed that miR-17 regulates SIK1 by directly binding to its 3ʹUTR in CRC cells.
It has been reported that decreased SIK1 expression is associated with poor prognosis in ovarian cancer, liver cancer, gastric adenocarcinoma, and pancreatic cancer.15–18,37,38 The underlying mechanism of SIK1 function in cancer is still unknown. Emerging evidence suggests that SIK1 may regulate cancer cell progression and metastasis by regulating the Wnt/β-catenin signaling pathway in HCC.15 The researchers found that SIK1 silencing may promote the metastasis of HCC, cell migration and lung metastasis, and that the loss of SIK1 may activate the Wnt/β-catenin signaling pathway, which mediates HCC cell proliferation and invasion. The Wnt/β-catenin signaling pathway may influence cellular differentiation and is associated with a variety of human cancers, including CRC.39,40 In the present study, we further investigated the prognostic value of SIK1 in CRC. Immunohistochemical analysis of 101 surgically resected CRC tissues revealed that SIK1 was significantly downregulated in tumor tissues in comparison to adjacent non-tumor tissues. This phenotype was further validated by qRT-PCR and Western blot analysis, which demonstrated lower SIK1 mRNA and protein expression, respectively, in tumor tissues. Additionally, lower expression of SIK1 protein in tissue samples from the enrolled CRC patients was significantly associated with tumor location, tumor differentiation, and TNM disease stage. In the analysis of overall survival, we also found that the expression of SIK1 may be an independent risk factor for predicting unfavorable clinical outcomes of CRC patients.
It is important to point out that a single microRNA may have multiple target genes, and a single gene may be targeted by multiple miRNAs. Studies have shown that HBP1 may be directly targeted by miR-17 in breast cancer.41 In pancreatic cancer, microRNA-203 promotes cell proliferation and migration by downregulating SIK1.17 In ovarian cancer, SIK1 was downregulated by microRNA-141.18 In our study, we identified that SIK1 may be a direct target of miR-17, and we hypothesized that miR-17 promotes CRC carcinogenesis by directly targeting SIK1. To our knowledge, this is the first report of SIK1 as a direct target of miR-17 in CRC. Based on previous studies and our current data, SIK1 may be a tumor suppressor gene and exert anti-tumor effects in HCC, pancreatic cancer, and CRC, which suggests that is may be a potential therapeutic target for human cancers. Therefore, a larger and more varied cohort of patient samples is required for future studies to more comprehensively assess the potential biological function of miR-17 and SIK1.
Conclusion
In summary, our study indicates that miR-17 expression is upregulated in colorectal cancer tissues and correlated with poor prognosis. Further functional investigation revealed that downregulation of miR-17 may promote tumor proliferation and invasion, through direct targeting of SIK1. Our results suggest that SIK1 and miR-17 may be potential prognostic biomarkers and novel potential treatment targets for patients with CRC.
Abbreviation list
CRC, colorectal cancer; SIK1, salt-inducible kinase 1; AMPK, AMP-activated protein kinase; RT-PCR, real-time polymerase chain reaction; BSA, bovine serum albumin; HR, hazard ratio; CI, confidence interval; HCC, hepatocellular carcinoma; miRNA, microRNA.
Acknowledgment
We are grateful to all the patients who enrolled in this study. This work was supported by grants from the Science and Technology Planning Project of Guangdong Province, China (No. 2013B021800161, 2014A070705012, 2016A02021512-8001, 2017A0300223006) and Science and Technology Project of Guangzhou, China (No. 201704020077).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
3. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
4. Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23(3):609–618. doi:10.1200/JCO.2005.01.086
5. Bjerkeset T, Morild I, Mork S, Soreide O. Tumor characteristics in colorectal cancer and their relationship to treatment and prognosis. Dis Colon Rectum. 1987;30(12):934–938.
6. Wang WS, Lin JK, Chiou TJ, et al. CA19-9 as the most significant prognostic indicator of metastatic colorectal cancer. Hepatogastroent-erology. 2002;49(43):160–164.
7. Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72(5–6):397–402. doi:10.1159/000113489
8. Pu J, Bai D, Yang X, Lu X, Xu L, Lu J. Adrenaline promotes cell proliferation and increases chemoresistance in colon cancer HT29 cells through induction of miR-155. Biochem Biophys Res Commun. 2012;428(2):210–215. doi:10.1016/j.bbrc.2012.09.126
9. Li ZW, Yang YM, Du LT, et al. Overexpression of miR-223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31(11):1–5. doi:10.1007/s12032-014-0256-5
10. Yang X, Hu Y, Liu Y, et al. C14orf28 downregulated by miR-519d contributes to oncogenicity and regulates apoptosis and EMT in colorectal cancer. Mol Cell Biochem. 2017;434(1–2):197–208. doi:10.1007/s11010-017-3049-2
11. Huang C, Yu M, Yao X. MicroRNA-17 and the prognosis of human carcinomas: a systematic review and meta-analysis. BMJ Open. 2018;8(5):e018070. doi:10.1136/bmjopen-2017-018070
12. Qian W, Li YX, Liu M, Li X, Hua T. MiR-17-5p targets TP53INP1 and regulates cell proliferation and apoptosis of cervical cancer cells. IUBMB Life. 2012;64(8):697–704. doi:10.1002/iub.1095
13. Selvik LKM, Rao S, Steigedal TS, et al. Salt-inducible kinase 1 (SIK1) is induced by gastrin and inhibits migration of gastric adenocarcinoma cells. PLoS One. 2014;9(11):e112485–e112485. doi:10.1371/journal.pone.0112485
14. Berdeaux R, Goebel N, Banaszynski L, et al. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat Med. 2007;13(5):597. doi:10.1038/nm1573
15. Qu C, He D, Lu X, et al. SIK1 silencing promotes HCC progression and WNT/β-catenin activation. J Hepatol. 2016. doi:10.1016/j.jhep.2016.01.005
16. Cheng H, Liu P, Wang ZC, et al. SIK1 couples LKB1 to p53-dependent anoikis and suppresses metastasis. Sci Signal. 2009;2(80):ra35. doi:10.1126/scisignal.2000369
17. Ren ZG, Dong SX, Han P, Qi J. miR-203 promotes proliferation, migration and invasion by degrading SIK1 in pancreatic cancer. Oncol Rep. 2016;35(3):1365–1374. doi:10.3892/or.2015.4534
18. Chen JL, Chen F, Zhang TT, Liu NF. Suppression of SIK1 by miR-141 in human ovarian cancer cell lines and tissues. Int J Mol Med. 2016;37(6):1601. doi:10.3892/ijmm.2016.2553
19. Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with image. J Biophotonics Int. 2004;11(5–6):36–42.
20. Rhodes DR, Kalyanasundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180.
21. Gaedcke J, Grade M, Jung K, et al. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosomes Cancer. 2010;49(11):1024–1034. doi:10.1002/gcc.20811
22. Kaiser S, Park YK, Franklin JL, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8(7):R131. doi:10.1186/gb-2007-8-5-r81
23. Skrzypczak M, Goryca K, Rubel T, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5(10):e13091. doi:10.1371/journal.pone.0013091
24. Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis. 2010;27(2):83–90. doi:10.1007/s10585-010-9305-4
25. Agarwal V, Bell GW, Nam JW, Bartel D. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005.
26. Dweep H, Gretz NJNM. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697.
27. Dweep H, Sticht C, Pandey P, Gretz N. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44(5):839–847. doi:10.1016/j.jbi.2011.05.002
28. Efron B. Logistic regression, survival analysis, and the Kaplan-Meier curve. J Am Stat Assoc. 1988;83(402):414–425. doi:10.1080/01621459.1988.10478612
29. Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther. 2010;10(8):748–757. doi:10.4161/cbt.10.8.13083
30. Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–9632. doi:10.1158/0008-5472.CAN-05-2352
31. Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26(21):8191–8201. doi:10.1128/MCB.00242-06
32. Lichner Z, Saleh C, Subramaniam V, Seivwright A, Prud’homme GJ, Yousef GM. miR-17 inhibition enhances the formation of kidney cancer spheres with stem cell/tumor initiating cell properties. Oncotarget. 2015;6(8):5567. doi:10.18632/oncotarget.v6i8
33. Zhong W, Chen S, Qin Y, et al. Doxycycline inhibits breast cancer EMT and metastasis through PAR-1/NF-κB/miR-17/E-cadherin pathway. Oncotarget. 2017;8(62):104855–104866. doi:10.18632/oncotarget.20418
34. Lekun F, Haoran L, Lei W, et al. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5(10):2974–2987.
35. Ma Y, Zhang P, Wang F, et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun. 2012;3(4):1291.
36. Maziere P, Enright AJJDDT. Prediction of microRNA targets. 2007;12(11):452–458. Drug Discov Today. doi:10.1016/j.drudis.2007.04.002
37. Yao YH, Cui Y, Qiu XN, et al. Attenuated LKB1-SIK1 signaling promotes epithelial-mesenchymal transition and radioresistance of non-small cell lung cancer cells. Chin J Cancer. 2016;35:50. doi:10.1186/s40880-016-0113-3
38. Selvik LK, Rao S, Steigedal TS, et al. Salt-inducible kinase 1 (SIK1) is induced by gastrin and inhibits migration of gastric adenocarcinoma cells. PLoS One. 2014;9(11):e112485. doi:10.1371/journal.pone.0112485
39. Jumpertz S, Hennes T, Asare Y, Schütz AK, Bernhagen J. CSN5/JAB1 suppresses the WNT inhibitor DKK1 in colorectal cancer cells. Cell Signal. 2017;34:38. doi:10.1016/j.cellsig.2017.02.013
40. Hlubek F, Brabletz T, Budczies J, Pfeiffer S, Jung A, Kirchner T. Heterogeneous expression of Wnt/β-catenin target genes within colorectal cancer. Int J Cancer. 2007;121(9):1941–1948. doi:10.1002/ijc.22916
41. Li H, Bian C, Liao L, Li J, Zhao RC. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat. 2011;126(3):565–575. doi:10.1007/s10549-010-0954-4
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.