Back to Journals » OncoTargets and Therapy » Volume 11
MicroRNA-150 suppresses triple-negative breast cancer metastasis through targeting HMGA2
Authors Tang W, Xu P, Wang H , Niu Z, Zhu D, Lin Q, Tang L, Ren L
Received 9 January 2018
Accepted for publication 21 February 2018
Published 24 April 2018 Volume 2018:11 Pages 2319—2332
DOI https://doi.org/10.2147/OTT.S161996
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tohru Yamada
Wentao Tang,1,* Pingping Xu,1,* Hong Wang,1,* Zhengchuan Niu,1 Dexiang Zhu,1 Qi Lin,1 Liming Tang,2 Li Ren1
1Department of General Surgery, Zhongshan Hospital, Fudan University, Shanghai, China; 2Department of General Surgery, Affiliated Changzhou No 2 People’s Hospital, Nanjing Medical University, Changzhou, China
*These authors contributed equally to this work
Background: Growing evidence suggests that miR-150 plays an inhibitory role in various types of cancer. However, the function and underlying mechanisms of miR-150 in triple-negative breast cancer (TNBC) remain unknown.
Patients and methods: miR-150 expression was detected by qRT-PCR and ISH in TNBC tumor and adjacent normal breast tissues. miR-150 function was analyzed by wound healing and transwell assay in vitro and mouse lung metastasis model in vivo. mRNA microarray, qRT-PCR, western blotting and luciferase assay were used to identify the target gene of miR-150. HMGA2 over-expression plasmid was co-transfected with miR-150 to study the role of miR-150 through regulating HMGA2.
Results: We found that miR-150 was down-regulated in TNBC tumor tissues compared to corresponding adjacent, normal breast tissues, and was correlated with decreased lymph-node metastasis. Ectopic expression of miR-150 suppressed TNBC cell migration in vitro and metastasis in vivo. Mechanistic study revealed that miR-150 down-regulates HMGA2 by directly targeting its mRNA. Moreover, the suppression of cell migration caused by miR-150 is relieved by over-expression of HMGA2, suggesting that miR-150 inhibits migration of TNBC cells by down-regulating HMGA2.
Conclusion: This work indicates that the miR-150/HMGA2 axis may serve as a treatment marker in TNBC.
Keywords: miR-150, HMGA2, triple-negative breast cancer, metastasis
Introduction
Breast cancer is the most common malignancy among women worldwide, with an annual rate of increase of 3.1%, and the second leading cause of cancer-related mortality in women.1,2 It is a highly heterogeneous disease and is made up of different subtypes. Based on different gene expression patterns, breast cancer can be classified into five subtypes: luminal A, luminal B, basal-like, ERBB2+, and normal breast-like.3 It can also be classified based on the presence or absence of receptors, such as estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER2). Triple-negative breast cancer (TNBC), without the expression of ER, PR, and HER2, is a special subtype and lacks the targets for anti-hormone and anti-HER2 therapies. Although TNBC accounts for only 10%–20% of all newly diagnosed breast cancers, it has been the focus of research owing to its aggressive clinical features.4,5 Patients with TNBC are more commonly diagnosed at a younger age (<50 years) and develop larger tumors, and have a higher chance of recurrence, distant metastasis, and death.6 Although TNBC patients are sensitive to chemotherapy, they are prone to medical treatment resistance.7 Therefore, there is an urgent need to screen and clarify new treatment targets for TNBC.
miRNAs are a class of endogenous small non-coding RNAs that regulate gene expression by targeting the 3′ non-coding area of mRNA. Increasing evidence shows that miRNAs regulate many cellular processes through down-regulating the expression of target genes.8 Nowadays, miRNAs have become attractive candidate biomarkers for cancer diagnosis and prognosis, and novel targets for cancer treatment because of their high stability and specific expression pattern during tumorigenesis and progression.9
As a hematopoiesis-related miRNA, miR-150 has been frequently studied in normal hematopoiesis and various types of hematopoietic malignancies.10 Through genome-wide miRNA expression profiling, miR-150 was identified as one of the most down-regulated miRNAs and a pivotal tumor suppressor gene of acute myeloid leukemia (AML).11 miR-150 was also reported to play an inhibitory role in the vast majority of solid tumors, including ovarian cancer, liver cancer, pancreatic cancer, colorectal cancer, and head and neck squamous cell carcinoma,12–16 although its role in lung cancer was not consistent among different studies.17–19 miR-150 has also been reported to be over-expressed in breast cancer tissues. High expression of miR-150 promotes breast cancer growth and reduces apoptosis through targeting the P2X7 receptor, indicating that it was an onco-miRNA in breast cancer.20 However, miRNA expression profiling of 165 TNBC samples and 59 normal controls demonstrated that miR-150 was down-regulated in TNBC samples.21 Moreover, miRNA global expression profiling from a well-annotated cohort with complete 10-year follow-up revealed that the level of miR-150 was independently associated with distant relapse-free survival of TNBC.22 These studies indicate that miR-150 may play an inhibitory role in TNBC.
HMGA2, a member of the high-mobility group protein family, is a chromatin remodeling factor which binds to AT-rich regions in DNA.23 HMGA2 is highly expressed during embryogenesis yet not in normal adult tissues.24 HMGA2 is over-expressed in a number of malignant tumors and high expression of HMGA2 is correlated with tumor staging and malignant phenotype.25–28 Studies have shown that HMGA2 is involved in the regulation of tumorigenesis, growth, epithelial–mesenchymal transition, and metastasis of breast cancer.29–32
In the present study, we found that miR-150 is less expressed in TNBC and negatively correlated with lymph-node metastasis. It inhibits TNBC metastasis through down-regulating the oncogene HMGA2.
Patients and methods
Human tissues and cell lines
In total, 10 pairs of TNBC tissues and adjacent normal breast tissues, and another 30 TNBC samples were collected at Changzhou No. 2 People’s Hospital between January 2015 and December 2015. All patients have received radical tumor resections without preoperative adjuvant chemotherapy and/or radiotherapy. Lumps of surgical samples were snap-frozen in liquid nitrogen and stored at −80°C for miRNA detection. The remaining tissues were embedded for immunohistochemistry and RNA in situ hybridization (ISH) study. This study was approved by the Clinical Research Ethics Committee of Changzhou No. 2 People’s Hospital. All clinical samples and data were obtained with written informed consent.
Both human TNBC cell lines (MDA-MB-231 and Hs578T) and non-TNBC cell lines (BT-474 and MCF-7) were purchased from the Chinese Academy of Sciences (Shanghai, China). Both of them were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) at 37°C and 5% CO2.
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Complementary DNA was synthesized using the PrimeScript RT-PCR Kit (TaKaRa, Tokyo, Japan). qRT-PCR was performed using SYBR® Premix Ex Taq™ (Takara) on an ABI 7500 system (Applied Biosystems, Carlsbad, CA, USA). Bulge-Loop miRNA primers were ordered from RiboBio (Guangzhou, China). U6 and GAPDH were used as the internal control for miRNA and mRNA detection, respectively. The sequence of primers for mRNA detection is displayed in Table S1.
ISH and immunohistochemistry
ISH was used to detect miR-150 expression in TNBC samples. An oligonucleotide probe complementary to miR-150 was digoxigenin labeled at the 5′-terminal end, and detected using an ISH detection kit (Boster, Wuhan, China) according to the manufacturer’s instructions. miR-150 staining was scored according to staining intensity and proportion. The intensity score was record as 0 (negative), 1 (weakly positive), 2 (moderately positive), or 3 (strongly positive). The proportion score used was 0 (0%), 1 (>0 to ≤25%), 2 (>25% to ≤50%), 3 (>50% to ≤75%), or 4 (>75%). The final score was calculated as proportion score × intensity score.
Immunohistochemistry was performed to investigate HMGA2 expression in TNBC samples. The staining score was evaluated using the same method described in ISH. miR-150 and HMGA2 staining were scored blindly by two pathologists.
Oligonucleotide transfection and lentivirus infection
miR-150 agomir and agomir control (ctrl) were ordered from RiboBio. The miRNA oligonucleotides were transfected by lipofectamine 2000 at a final concentration of 50 nmol/L (Invitrogen, Carlsbad, CA, USA).
The lentivirus over-expression human miR-150 and negative control were purchased from GeneChem (Shanghai, China). The primer sequence for miR-150 was: forward 5′-GAGGATCCCCGGGTACCGGTCTGGCAGGAACCCCCGCCCT-3′ and reverse 5′-CACACATTCCACAGGCTAGTAAAAGCCGCAGCAGAGATG-3′. For infection, the virus was supplemented with 8 μg/mL of polybrene (Sigma, St Louis, MO, USA). After infection for 72 h, 2 μg/mL puromycin (Sigma) was added to the cells, which underwent selection for 1 week.
Wound healing assay
After miRNA transfection for 24 h, MDA-MB-231 and Hs578T cells were harvested and reseeded into 96-well culture plates at the concentration of 3 × 104/well and 2 × 104/well, respectively. After 16 h incubation, when cells had reached more than 90% convergence, the wound was performed using a 96-pin wounding replicator (V&P Scientific, San Diego, CA, USA), washed twice in sterile PBS, replaced with serum-free medium, and maintained in an incubator. Cells were photographed after 0, 8, and 24 h, and the width of the wound was recorded as the wound distance. The wound healing distance was calculated by subtracting the value of the wound distance at 0 h from the value at 8 h or 24 h after scratching.
Transwell assay
The cell migration ability was analyzed by a transwell assay, which was performed in 24-well transwell chambers (Corning, NY, USA). After 24 h transfection, MDA-MB-231 cells (5 × 104) and Hs578T cells (3 × 104) in 200 μL serum-free medium were reseeded into the upper chamber. The lower chamber was filled with 600 μL medium with 10% FBS. After incubating for 12 h, cells on the inner membrane were removed. The outer membrane was fixed with 4% paraformaldehyde and stained with 0.1% crystal violet solution. The cells were observed under a microscope and five fields were randomly selected to be photographed.
In vivo models
Four-week-old male Nu/Nu mice (SLAC, Shanghai, China) were kept under specific pathogen-free conditions. First, 100 μL MDA-MB-231 cells (2 × 107/mL) over-expressing miR-150 or miR-ctrl were slowly injected into the tail vein of mice (five mice in each group). At both 6 and 8 weeks, mice were anesthetized, and luciferase activity was evaluated under a small animal live imaging system (PerkinElmer, Waltham, MA, USA). After 8 weeks, mice were killed, and the number of metastatic nodules in the lung of each mouse was calculated. This study was approved by the Animal Ethics Committee of Zhongshan hospital and complied with the Guide for the Care and Use of Laboratory Animals.
mRNA microarray analysis
Total RNAs from MDA-MB-231 cells transfected with miR-150 or miR-ctrl were reverse transcribed into cDNA, labeled with biotin using the GeneChip 3′IVT Express Kit (Affymetrix, Santa Clara, CA, USA), and then hybridized to a PrimeView Human Gene Expression Array (Applied Biosystems, Carlsbad, CA, USA). After hybridization, the array was washed and stained using Affymetrix GeneChip Wash and Stain Kit on a GeneChip Fluidics Station 450. Finally, the array was scanned through a GeneChip Scanner 3000.
Western blotting
Whole protein extracts were lysed and run on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and transferred to a polyvinylidene fluoride membrane (BioRad, Hercules, CA, USA). After blocking, the membranes were incubated with the following primary antibodies: anti-HMGA2 (1:1,000), anti-LCN2 (1:1,000), and anti-GAPDH (1:1,000) (Cell Signaling Technology, Danvers, MA, USA), and anti-MSI2 (1:1,000), anti-LOXL2 (1:1,000), and anti-PBX1 (1:1,000) (GeneTex, Irvine, CA, USA). After washing with Tris-buffered saline containing Tween-20 (TBST) and incubation with secondary antibodies, signals were developed using an enhanced chemiluminescence kit (Pierce, Waltham, MA, USA).
Vector construction
HMGA2 cDNA was amplified from the total cDNA of Hs578T cells and cloned into the pcDNA3.1(+) vector.
The 3′ untranslated region (UTR) of HMGA2 was amplified from human genomic DNA and cloned into the pGL3 vector (Promega, Madison, WI, USA). Mutation of the miR-150 target site in the 3′UTR of HMGA2 was performed using a site-directed mutagenesis kit (Takara). The primer sequences for pcDNA3.1-HMGA2, HMGA2 3′UTR, wild-type (WT), and mutant (Mut) plasmids are listed in Table S2.
Luciferase assay
HEK293T cells (1 × 105) were cultured in 24-well plates, then transfected with 25 nmol miRNA, 20 ng Renil, and 200 ng WT or Mut reporter plasmid using lipofectamine 2000. After 24 h transfection, the luciferase activities of firefly and Renilla were measured by a dual-luciferase reporter assay system (Promega, Madison, WI, USA).
Statistical analysis
Data are expressed as mean ± SD. Statistical differences between groups were calculated using the Student’s t-test. The statistical correlation between miR-150 and HMGA2 expression was determined by the Pearson χ2-test.
Results
miR-150 is frequently down-regulated in TNBC
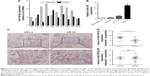
miR-150 expression was determined in 10 TNBC tumor samples and corresponding adjacent normal breast gland tissues by qRT-PCR. Lower miR-150 expression was found in seven out of 10 TNBC tumor samples (Figure 1A). Average miR-150 expression was approximately 1.6-fold lower in TNBC tumor specimens than in corresponding adjacent normal tissues. miR-150 was also expressed at a lower level in TNBC cell lines than in non-TNBC cell lines (Figure 1B). ISH analysis of another 30 TNBC tumor samples showed that the level of miR-150 was significantly lower in patients with lymph-node metastasis (p<0.05) (Figure 1C and D), but unrelated to tumor size (Figure 1E). These data suggest that miR-150 may negatively regulate TNBC cell metastasis.
miR-150 inhibits cell migration in vitro
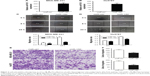
Two TNBC cell lines, MDA-MB-231 and Hs578T, were transfected with miR-150 agomir or miR-ctrl agomir. The efficiency of miR-150 transfection was confirmed by qRT-PCR analysis (Figure 1A and B). Although the migration ability of the cells in the two groups does not show a clear difference 8 h after scratching in MDA-MB-231 cells, the wound is much wider in the miR-150 group 24 h after scratching, demonstrating that miR-150 significantly inhibits the migration of MDA-MB-231 cells (Figure 2C). A similar phenotype was observed in Hs578T cells. Only 8 h after scratching, the inhibition of cell migration mediated by miR-150 is already visible (Figure 2D). The role of miR-150 in suppressing cell migration of MDA-MB-231 and Hs578T cells was further confirmed by transwell assay (Figure 2E and F).
miR-150 suppresses cell metastasis in vivo
Two stable cell lines, MDA-MB-231/miR-150 and MDA-MB-231/miR-ctrl, were established using lentivirus. These two stable cell lines were injected into the tail vein of mice (five mice in each group), and the ability of cancer metastasis was judged by analyzing the luciferase signal of lung metastasis 6 and 8 weeks after cell injection. In both cases, the luciferase intensity is significantly lower in miR-150 group, suggesting that it inhibits metastasis of TNBC in vivo (Figure 3A and B). The pictures of the whole lung and the results of H&E staining show that fewer metastatic nodules are formed in the lung from the miR-150 group (Figure 3C–E), further supporting the conclusion that miR-150 suppressed lung metastasis of MDA-MB-231 cells.
HMGA2 is a direct target of miR-150
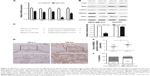
miRNAs function via base-pairing with complementary sequences within mRNA molecules and down-regulating the mRNA level. To identify potential mRNA targeted by miR-150, mRNA microarray was used to profile the total mRNA level of control or miR-150 over-expressing MDA-MB-231 cells. Compared with the control group, 318 mRNAs were down-regulated with a fold-change of more than 1.5, which are the potential targets of miR-150. The list was further narrowed down to 19 mRNAs by sequence-dependent prediction using both TargetScan and miRWalk2.0 software (Table S3). Among these mRNAs, the 10 most down-regulated ones were further analyzed by qRT-PCR, and the result shows that only five out of 10 mRNAs are down-regulated by miR-150 (lcn2, msi2, loxl2, hmga2, and pbx1) (Figure 4A), whereas the other five mRNAs are unaffected (data not shown). To further shorten the list, Western blotting was performed to analyze the protein level of these five genes. Only the protein level of HMGA2 was down-regulated in both cell lines, suggesting that this gene is the most likely target of miR-150 (Figure 4B). Down-regulation of MSI2 and PBX1 was only observed in MDA-MB-231 cells, indicating that the regulation of these two genes has cell-line specificity. No difference was observed in LCN2 and LOXL2 protein level (Figure 4B). Consistent with the idea that HMGA2 is a direct target of miR-150, miR-150 significantly reduced the luciferase activity of the WT reporter construct but had no effect on the mutant reporter construct, in which the sequence complementing miR-150 was mutated (Figure 4C). In addition, IHC staining of HMGA2 in the same 30 TNBC tumor samples demonstrated that HMGA2 was highly expressed in TNBC patients with lymph-node metastasis (Figure 4D and E), and the result of the Pearson χ2-test shows that HMGA2 and miR-150 expression were negatively correlated in TNBC (Figure 4F). Altogether, these data indicate that miR-150 directly targets HMGA2 and down-regulates its mRNA and protein level.
HMGA2 up-regulation partially relieves miR-150 mediated suppression of cell migration
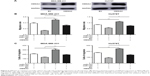
To determine whether miR-150 suppresses cancer cell migration by down-regulating oncogene HMGA2, vector over-expressing HMGA2 was constructed and transfected into MDA-MB-231 and Hs578T cells. Over-expression of HMGA2 was confirmed by Western blotting (Figure 5A). miR-150-mediated suppression of cell migration was partially rescued by HMGA2 in both MDA-MB-231 and Hs578T cells (Figure 5B). This observation was further confirmed by transwell assay (Figure 5C). Together, these data indicate that miR-50 suppresses cell migration by down-regulating HMGA2.
Discussion
Although aberrant expression of miRNAs, including miR-21, mir-26, miR-148a, miR-206, miR-200b, miR-455-3p, and miR-655, has been reported in TNBC metastasis,33–39 novel miRNAs involved in the regulation of TNBC metastasis still remain to be explored. In the present study, miR-150 was found to be less expressed in TNBC tissues compared to the corresponding adjacent normal breast tissues, and decreased miR-150 level is correlated with increased lymph-node metastasis. miR-150 over-expression suppressed TNBC cells migration in vitro and lung metastasis in vivo. Moreover, HMGA2 was identified as a novel target of miR-150, and HMGA2 over-expression partially relieved the suppression of cell migration caused by miR-150. These observations indicate that miR-150 play an inhibitory role in TNBC metastasis through down-regulating HMGA2.
Although miR-150 is aberrantly down-regulated in most hematopoietic malignancies and solid tumors,10,14,15 the underlying mechanism remained largely unknown. Jiang and colleagues found that MLL-fusion proteins transactivated miR-150 through directly binding to the promoter region, but inhibited the maturation process of miR-150 through the MYC/LIN28 axis.11 It was also reported that the transcription of miR-150 was directly activated by the WNT/β-catenin pathway in colorectal cancer,40 but the expression of miR-150 was reduced compared with paired non-cancerous tissue.16 Since MYC was a direct target of the WNT pathway, the MYC/LIN28 axis may also play an import role in the down-regulation of miR-150 in colorectal cancer. In the present study, miR-150 was found to be less expressed in TNBC and negatively associated with lymph-node metastasis. The underlying mechanism may also be linked to MYC/LIN28, since an aberrant Wnt pathway was identified as participating in TNBC tumorigenesis and metastasis.41–43
Some studies have reported an inhibitory role of miR-150 in most solid tumors. Low expression of miR-150 was associated with a poor prognosis in patients with esophageal squamous cell carcinoma, epithelial ovarian cancer, and colorectal cancer, and over-expression of miR-150 inhibited epithelial mesenchymal transition through down-regulating ZEB-1 and GLI1.16,44–46 A similar role of miR-150 was also identified in liver cancer and non-small-cell lung cancer.47,48 In the present study, miR-150 was found to inhibit TNBC cell migration in vitro and metastasis in vivo, implying that miR-150 also played an inhibitory in TNBC. This result is consistent with a previous study showing that high miR-150 expression was associated with better distant relapse-free survival of TNBC patients.22 Therefore, miR-150 serves as a potential treatment target for various kinds of cancers.
Previous studies have shown that high expression of HMGA2 is critical for the pathogenesis and progression of a large number of tumors.26,29,49 It was proposed that the regulation of HMGA2 expression was involved in both transcriptional and post-transcriptional regulation in breast cancer. As for transcriptional regulation, it was reported that ZBRK1, BRCA1, and CtIP form a complex that coordinately inhibits HMGA2 expression via binding to HMGA2 promoter.29 This regulation may also serve as a mechanism for BRCA1 deficiency-induced breast tumorigenesis. In addition, altered HMGA2 protein level in tumors is not always correlated with an increased amount of corresponding mRNA, which indicates a post-transcriptional regulation mediated by regulatory elements within the 3′UTR.50 A good example is Let-7, a well-known tumor suppressor, which down-regulates HMGA2 via a post-transcriptional mechanism.51 miR-33b, miR-185, and miR-211 were also reported to regulate HMGA2 expression in breast cancer by targeting its mRNA.52–54 Here, we showed that miR-150 directly targets HMGA2, which is a novel miRNA-mediated modulation of HMGA2 in TNBC. Besides the above mechanisms, HMGA2 can also be regulated by the WNT pathway in TNBC.55,56
Conclusion
This study demonstrates that miR-150 is down-expressed in TNBC samples and negatively associated with lymph-node metastasis of TNBC patients. In addition, we found that miR-150 suppresses TNBC cell migration and metastasis by directly targeting HMGA2. This work indicates that the miR-150/HMGA2 axis may serve as a potential treatment target in TNBC.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (81402341).
Disclosure
The authors report no conflicts of interest in this work.
References
DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448. | ||
Forouzanfar MH, Foreman KJ, Delossantos AM, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378(9801):1461–1484. | ||
Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. | ||
Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15 Pt 1):4429–4434. | ||
Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109(9):1721–1728. | ||
Podo F, Buydens LM, Degani H, et al. Triple-negative breast cancer: present challenges and new perspectives. Mol Oncol. 2010;4(3):209–229. | ||
Metzger-Filho O, Tutt A, de Azambuja E, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30(15):1879–1887. | ||
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. | ||
Andorfer CA, Necela BM, Thompson EA, Perez EA. MicroRNA signatures: clinical biomarkers for the diagnosis and treatment of breast cancer. Trends Mol Med. 2011;17(6):313–319. | ||
He Y, Jiang X, Chen J. The role of miR-150 in normal and malignant hematopoiesis. Oncogene. 2014;33(30):3887–3893. | ||
Jiang X, Huang H, Li Z, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22(4):524–535. | ||
Kim TH, Jeong JY, Park JY, et al. miR-150 enhances apoptotic and anti-tumor effects of paclitaxel in paclitaxel-resistant ovarian cancer cells by targeting Notch3. Oncotarget. 2017;8(42):72788–72800. | ||
Sun W, Zhang Z, Wang J, et al. MicroRNA-150 suppresses cell proliferation and metastasis in hepatocellular carcinoma by inhibiting the GAB1-ERK axis. Oncotarget. 2016;7(10):11595–11608. | ||
Yang K, He M, Cai Z, et al. A decrease in miR-150 regulates the malignancy of pancreatic cancer by targeting c-Myb and MUC4. Pancreas. 2015;44(3):370–379. | ||
Koshizuka K, Nohata N, Hanazawa T, et al. Deep sequencing-based microRNA expression signatures in head and neck squamous cell carcinoma: dual strands of pre-miR-150 as antitumor miRNAs. Oncotarget. 2017;8(18):30288–30304. | ||
Ma Y, Zhang P, Wang F, et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61(10):1447–1453. | ||
Cao M, Hou D, Liang H, et al. miR-150 promotes the proliferation and migration of lung cancer cells by targeting SRC kinase signalling inhibitor 1. Eur J Cancer. 2014;50(5):1013–1024. | ||
Li H, Ouyang R, Wang Z, et al. MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci Rep. 2016;6:39001. | ||
Lu W, Zhang H, Niu Y, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16(1):118. | ||
Huang S, Chen Y, Wu W, et al. miR-150 promotes human breast cancer growth and malignant behavior by targeting the pro-apoptotic purinergic P2X7 receptor. PLoS One. 2013;8(12):e80707. | ||
Cascione L, Gasparini P, Lovat F, et al. Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS One. 2013;8(2):e55910. | ||
Buffa FM, Camps C, Winchester L, et al. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71(17):5635–5645. | ||
Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7(12):899–910. | ||
Fedele M, Visone R, De Martino I, et al. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9(6):459–471. | ||
Rogalla P, Drechsler K, Kazmierczak B, Rippe V, Bonk U, Bullerdiek J. Expression of HMGI-C, a member of the high mobility group protein family, in a subset of breast cancers: relationship to histologic grade. Mol Carcinog. 1997;19(3):153–156. | ||
Abe N, Watanabe T, Suzuki Y, et al. An increased high-mobility group A2 expression level is associated with malignant phenotype in pancreatic exocrine tissue. Br J Cancer. 2003;89(11):2104–2109. | ||
Meyer B, Loeschke S, Schultze A, et al. HMGA2 overexpression in non-small cell lung cancer. Mol Carcinog. 2007;46(7):503–511. | ||
Huang ML, Chen CC, Chang LC. Gene expressions of HMGI-C and HMGI(Y) are associated with stage and metastasis in colorectal cancer. Int J Colorectal Dis. 2009;24(11):1281–1286. | ||
Ahmed KM, Tsai CY, Lee WH. Derepression of HMGA2 via removal of ZBRK1/BRCA1/CtIP complex enhances mammary tumorigenesis. J Biol Chem. 2010;285(7):4464–4471. | ||
Sun M, Song CX, Huang H, et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci U S A. 2013;110(24):9920–9925. | ||
Yang E, Cisowski J, Nguyen N, et al. Dysregulated protease activated receptor 1 (PAR1) promotes metastatic phenotype in breast cancer through HMGA2. Oncogene. 2016;35(12):1529–1540. | ||
Tan EJ, Kahata K, Idas O, Thuault S, Heldin CH, Moustakas A. The high mobility group A2 protein epigenetically silences the Cdh1 gene during epithelial-to-mesenchymal transition. Nucleic Acids Res. 2015;43(1):162–178. | ||
Fang H, Xie J, Zhang M, Zhao Z, Wan Y, Yao Y. miRNA-21 promotes proliferation and invasion of triple-negative breast cancer cells through targeting PTEN. Am J Transl Res. 2017;9(3):953–961. | ||
Liu P, Tang H, Chen B, et al. miR-26a suppresses tumour proliferation and metastasis by targeting metadherin in triple negative breast cancer. Cancer Lett. 2015;357(1):384–392. | ||
Xu X, Zhang Y, Jasper J, et al. MiR-148a functions to suppress metastasis and serves as a prognostic indicator in triple-negative breast cancer. Oncotarget. 2016;7(15):20381–20394. | ||
Liang Z, Bian X, Shim H. Downregulation of microRNA-206 promotes invasion and angiogenesis of triple negative breast cancer. Biochem Biophys Res Commun. 2016;477(3):461–466. | ||
Humphries B, Wang Z, Oom AL, et al. MicroRNA-200b targets protein kinase Cα and suppresses triple-negative breast cancer metastasis. Carcinogenesis. 2014;35(10):2254–2263. | ||
Li Z, Meng Q, Pan A, et al. MicroRNA-455-3p promotes invasion and migration in triple negative breast cancer by targeting tumor suppressor EI24. Oncotarget. 2017;8(12):19455–19466. | ||
Lv ZD, Kong B, Liu XP, et al. miR-655 suppresses epithelial-to-mesenchymal transition by targeting Prrx1 in triple-negative breast cancer. J Cell Mol Med. 2016;20(5):864–873. | ||
Guo YH, Wang LQ, Li B, et al. Wnt/β-catenin pathway transactivates microRNA-150 that promotes EMT of colorectal cancer cells by suppressing CREB signaling. Oncotarget. 2016;7(27):42513–42526. | ||
Xu J, Prosperi JR, Choudhury N, Olopade OI, Goss KH. β-Catenin is required for the tumorigenic behavior of triple-negative breast cancer cells. PLoS One. 2015;10(2):e0117097. | ||
Mohammed MK, Shao C, Wang J, et al. Wnt/β-catenin signaling plays an ever-expanding role in stem cell self-renewal, tumorigenesis and cancer chemoresistance. Genes Dis. 2016;3(1):11–40. | ||
Dey N, Barwick BG, Moreno CS, et al. Wnt signaling in triple negative breast cancer is associated with metastasis. BMC Cancer. 2013;13:537. | ||
Yokobori T, Suzuki S, Tanaka N, et al. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013;104(1):48–54. | ||
Jin M, Yang Z, Ye W, Xu H, Hua X. MicroRNA-150 predicts a favorable prognosis in patients with epithelial ovarian cancer, and inhibits cell invasion and metastasis by suppressing transcriptional repressor ZEB1. PLoS One. 2014;9(8):e103965. | ||
Fan H, Liu X, Zheng WW, Zhuang ZH, Wang CD. MiR-150 alleviates EMT and cell invasion of colorectal cancer through targeting Gli1. Eur Rev Med Pharmacol Sci. 2017;21(21):4853–4859. | ||
Li T, Xie J, Shen C, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75(15):3181–3191. | ||
Zhang HY, Zheng FS, Yang W, Lu JB. The long non-coding RNA MIAT regulates zinc finger E-box binding homeobox 1 expression by sponging miR-150 and promoteing cell invasion in non-small-cell lung cancer. Gene. 2017;633:61–65. | ||
Sun J, Sun B, Zhu D, et al. HMGA2 regulates CD44 expression to promote gastric cancer cell motility and sphere formation. Am J Cancer Res. 2017;7(2):260–274. | ||
Borrmann L, Wilkening S, Bullerdiek J. The expression of HMGA genes is regulated by their 3′UTR. Oncogene. 2001;20(33):4537–4541. | ||
Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. | ||
Lin Y, Liu AY, Fan C, et al. MicroRNA-33b inhibits breast cancer metastasis by targeting HMGA2, SALL4 and Twist1. Sci Rep. 2015;5:9995. | ||
Zou Q, Wu H, Fu F, Yi W, Pei L, Zhou M. RKIP suppresses the proliferation and metastasis of breast cancer cell lines through up-regulation of miR-185 targeting HMGA2. Arch Biochem Biophys. 2016;610:25–32. | ||
Li X, Wang S, Li Z, et al. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int J Biol Macromol. 2017;105(Pt 1):346–353. | ||
Wend P, Runke S, Wend K, et al. WNT10B/β-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol Med. 2013;5(2):264–279. | ||
Fatima I, El-Ayachi I, Taotao L, et al. The natural compound Jatrophone interferes with Wnt/β-catenin signaling and inhibits proliferation and EMT in human triple-negative breast cancer. PloS One. 2017;12(12):e0189864. |
Supplementary materials
  | Table S1 Specific real-time quantitative polymerase chain reaction primers |
  | Table S2 Polymerase chain reaction primers for constructs |
  | Table S3 Potential targets predicted by both TargetScan and miRWalk2.0 software |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.