Back to Archived Journals » Advanced Health Care Technologies » Volume 3
Microfluidics technology: future prospects for molecular diagnostics
Authors Lo RC
Received 29 June 2016
Accepted for publication 13 January 2017
Published 23 February 2017 Volume 2017:3 Pages 3—17
DOI https://doi.org/10.2147/AHCT.S94024
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Gozde Durmus
Roger C Lo
Department of Chemical Engineering, California State University, Long Beach, CA, USA
Abstract: Molecular diagnostic tests have been widely used to detect and quantify biomarkers, such as nucleic acids and proteins, associated with specific health conditions or diseases to obtain the critical information for the health care providers and patients to make correct medical decisions. However, existing diagnostic instruments usually require costly reagents, long analysis time, established lab infrastructure, and trained professionals to operate, which limits their availability for large-scale screening applications in developed countries and primary care in developing countries. There is a strong demand for robust, cost-effective, and simple-to-operate instruments for molecular diagnostics. The features of microfluidics, such as short analysis time, reduction in fabrication costs, and low sample/reagent consumption, make it a natural fit for the development of new diagnostic instruments. Herein, selected work is highlighted to provide a snapshot of microfluidic devices developed for molecular diagnostics in the past 5 years, specifically focusing on their applications for the detection of agents clinically relevant to cancers, cardiac conditions, and infectious diseases, and an outlook on how microfluidics technology can be further advanced for applications in this area.
Keywords: microfluidics, micro total analysis systems (µTAS), lab-on-a-chip, molecular diagnostics, point-of-care testing
Introduction
Microfluidics includes the science of studying and controlling the behaviors of fluids and particles at microscale (typically tens to hundreds of micrometers) and the technology of developing methods and devices that can precisely and effectively perform such tasks. For over two decades, microfluidics has found broad applications at the interface of biology, chemistry, engineering, and medicine, such as biodefense, genomics, proteomics, pharmaceuticals, and tissue engineering.1 Microfluidics has been widely employed to miniaturize analytical methods and biological/chemical processes because of its benefits such as portability, high throughput by parallelization, significant increase in sample process speed, reduction in fabrication costs and sample/reagent volumes (10−6–10−18 L), and enhanced system performance and functionality through the integration of various components onto individual devices.2,3 These applications are usually referred to as micro total analysis systems4 or lab-on-a-chip.5
The beginning of microfluidics can be traced back to the theoretical work developed for gas chromatography (GC)6 and liquid chromatography (LC) in the 1950s,7 which indicates that the separation performance can be improved by reducing the sizes of the column diameters and packing particles. This introduced the use of fused silica capillaries with diameters in micrometers in those systems. Later, capillary electrophoresis (CE) was developed to replace the conventional slab gel electrophoresis for the separation of charged biomolecules. The high surface-to-volume ratio of capillaries allowed fast heat dissipation to mitigate the Joule heating caused by the applied electric field. Therefore, electrophoretic separations could be performed at much higher electric fields, which led to extremely high speeds and resolutions.8 Although the benefits of miniaturizing analytical methods were identified, there was no suitable platform for further development of miniaturized analytical systems until the rise of the microelectronics industry. Its silicon-based micromachining processes (including photolithography, etching, and bonding steps) were first developed to fabricate microelectromechanical systems (MEMS)9 and proved to be a suitable platform for miniaturizing analytical methods, as supported by the success of the first GC air analyzer fabricated on a silicon wafer developed by Terry et al in 1979 (Figure 1A).10,11 This merge of analytical methods and miniaturization through microfabrication led to the creation of microfluidics. While the efforts of miniaturizing GC and LC columns continued (Figure 1B),12–16 the major success of miniaturizing analytical methods came from microfluidic separation chips based on electrophoresis due to their simpler design of applying an electric potential over a microchannel instead of the high-pressure source required by their GC/LC counterparts. In early 1990s, Manz et al demonstrated the potential of microfluidics to address the issues of conventional analytical methods with their chip-based LC and CE systems with integrated monitoring components.12,17–19 This marked the dawn of the modern microfluidics technology.
  | Figure 1 Miniaturized analytical systems. Notes: (A) Gas chromatographic air analyzer on a planar silicon wafer. It was composed of a sample injection system, a 1.5 m circular-spiral column (200×30 µm), and a thermal conductivity detector. (B) Liquid chromatography chip fabricated by Manz et al12 at Hitachi Central Research Laboratories. The silicon/glass chip included an open-tubular separation column (6 µm×2 µm×150 mm) and an integrated platinum electrode detector. Adapted from de Mello AJ. FOCUS On-chip chromatography: the last twenty years. Lab Chip. 2002;2(3):48N–54N. With permission from The Royal Society of Chemistry. © The Royal Society of Chemistry 2002. http://dx.doi.org/10.1039/b207266c.11 |
The choice of materials and fabrication methods of microfluidic devices are determined by their final applications. The first generation of microfluidic devices was fabricated with silicon and glass using the well-established micromachining processes from the microelectronics industry. Driven by new applications and performance requirements, new materials have been used to fabricate microfluidic devices such as ceramic,20 steel,21 Teflon,22 and paper.23 For academic research, polydimethylsiloxane (PDMS) is the most common material for microfluidic device fabrication because of its many advantages such as optical transparency, biocompatibility, elasticity, and a simple fabrication process (soft lithography) based on molding approaches.24–26 In spite of all the advantages, the commercialization of PDMS chips has limited success due to two major roadblocks. First, the multiple-step fabrication (including curing, access port punching, and assembly) is a mostly manual process that is difficult to be fully automated. As a result, the fabrication cost is high and the production is hard to scale up. Second, the nonstandard user interfaces (usually punched access ports) on PDMS chips are hard to make reliable, leak-free connections, as compared to industrial standards, for example, Luer-Lok and barbed connectors. To address these issues, thermoplastics, such as cyclic olefin copolymers and polystyrene,27 have been used as alternative materials for microfluidic chips. They can be processed by thermoforming,28 hot embossing,29 and injection molding,30 which makes it possible to mass produce microfluidic chips with integrated connectors. However, injection molding is not an ideal fabrication process for the science community because for high throughput and resolution, it is usually associated with expensive equipment and long turnaround times for the mold fabrication.
Additive manufacturing (commonly known as three-dimensional [3D] printing) was developed in the 1980s31 and had been of limited use for the general public until the expiration of a key patent in 2009.32 Since 2010, this technology has drawn more and more attention from the science community as a new tool for microfluidic chip fabrication, as reflected on the increased publications. The advantages of 3D printing for microfluidic chip fabrication are as follows. 1) One-step fabrication: a 3D model of a microfluidic chip can be created with any computer-aided design software, such as AutoCAD and SOLIDWORKS, and then sent to a 3D printer to build the chip through layer-by-layer construction. This fabrication process does not require cleanroom facilities like that for PDMS chips and can be fully automated. In addition, it allows multiple materials to be used and microstructures that are not compatible with conventional fabrication processes. 2) Capability of rapid prototyping: depending on the design complexity, the fabrication of a microfluidic chip can be completed in tens of minutes to a few hours from the digital data to an actual object. This allows researchers to take the “fail fast and often” strategy to significantly shorten the development cycle of microfluidic chips for desired applications and also makes it easier to commercialize viable designs into products. As of now, 3D printing cannot compete with conventional fabrication processes for microfluidic chips in terms of resolution, shape conformity, surface quality, and material availability, but it has shown great promise for future microfluidic chip fabrication. More information on 3D printing is available in several reviews and the references cited therein.33–37
This paper is intended to provide a snapshot of microfluidic devices developed for molecular diagnostics in the past 5 years, specifically focusing on their applications for the detection of agents clinically relevant to cancers, cardiac conditions, and infectious diseases, and an outlook on how microfluidics technology can be further advanced for applications in this area. Due to the diversity in microfluidics technology research and space limit, the author does not aim to provide a thorough and comprehensive review, but to give nonspecialist readers a general idea of how microfluidics technology can be integrated with conventional biomolecular techniques for enhanced performance and portability of molecular diagnostic tests for more effective health care delivery. For interested readers, more details on microfluidics technology for biological/biomedical research and applications can be found in several publications and the references cited therein.38–41
The need for improved molecular diagnostics
Molecular diagnostics is a collection of diagnostic tests used to assess an individual’s health at the molecular level by detecting and analyzing biological molecules such as nucleic acids (DNA and RNA) and proteins. These biological molecules serve as “biomarkers” (biological markers), which are indicators of biological states that can provide information to assist diagnosis, monitor disease progress, assess treatment responses, and identify the risk factors for disease prevention.42 For effective health care delivery, early and accurate detection is critical and can increase survivability, improve disease management and treatment outcomes, and ultimately lead to preventive and personalized medicine. Compared to conventional culture-based tests, molecular diagnostic tests have a higher sensitivity and specificity and a much shorter turnaround time. For example, the turnaround time for the diagnosis of sepsis is <10 h for molecular diagnostic tests instead of at least 72 h for culture-based tests (even longer for slow-growing pathogens such as yeasts and anaerobes).43,44 In clinical diagnosis, nucleic acids (DNA and RNA) and proteins are the mostly used molecular biomarkers in several general areas, including oncology, genetic disease screening, infectious diseases, coagulation, pharmacogenomics, and human leukocyte antigen typing.45 There is a strong demand for new molecular diagnostic tests with improved sensitivities, increased accuracy, shorter turnaround time, and low cost. Because of its advantages, microfluidics technology holds the promise for developing such molecular diagnostic tests through miniaturizing molecular diagnostic techniques onto microfluidic devices, as demonstrated by the work described in the following sections.
Microfluidic devices for nucleic acid-based tests
Polymerase chain reaction (PCR), gel electrophoresis, and hybridization are the techniques widely employed to detect and quantify nucleic acids for the identification of genomic biomarkers, such as mutations, polymorphisms, and epigenetic modifications.46–48
PCR is a technique for amplifying DNA fragments, and each reaction requires a DNA template, complementary primers, DNA polymerase (copying enzyme), deoxynucleotide triphosphates (building blocks for copies), and buffer. The mixture goes through thermal cycling to make DNA copies. Each PCR cycle contains three steps: denaturing (90°C–95°C), annealing (50°C–65°C), and polymerization (72°C–80°C). During denaturing, the double-stranded DNA template is separated into single strands. During annealing, the primers bind to the complementary bases on the single-stranded DNA (ssDNA). During polymerization, DNA polymerase reads the template and matches it with the corresponding deoxynucleotide triphosphates to assemble a complementary strand. The whole process is typically repeated 30–40 times to generate millions of identical copies of the original target DNA for analysis and takes about 2 h on a conventional system. For RNAs, reverse transcriptase PCR is used to amplify a target sequence. A double-stranded complementary DNA is first synthesized from the target RNA sequence with an enzyme, reverse transcriptase, from a retrovirus and then goes through the same PCR process as described earlier.
Gel electrophoresis is a technique for size-selective fractionation of charged target molecules, such as DNA and proteins, in a sieving matrix (“gel”; usually agarose and polyacrylamide) under the influence of an applied electric field. A typical gel electrophoresis run is carried out by injecting a mixture with target molecules of different sizes at the inlet of a gel-filled separation channel, after which the target molecules are transported through the gel under the action of an applied electric field. The migration velocities of the target molecules depend on their sizes, so the mixture separates into zones containing like-sized target molecules that are subsequently detected at a fixed downstream location. These zones provide the information on size differences of the target molecules that have diagnostic relevance. Depending on the sample size and resolution, it takes 45 min to 8 h to complete a separation run on a conventional system.
Hybridization is a technique based on the process where fluorescence-labeled, ssDNA or single-stranded RNA molecules serve as probes to specifically attach to those with complementary sequences, which can be used to obtain sequence-specific genetic information for diagnostic applications. There are three types of probes commonly used for diagnostics: 1) painting probes: they attach to overlapping sequences on a chromosome to apply chosen fluorescence colors; 2) centromeric probes: they target the centromeric region of a specific chromosome to help determine the number of copies of that specific chromosome; and 3) allele-specific probes: they attach to a specific allele sequence that is relevant to diagnostics. Here, we review selected microfluidic devices developed for these techniques.
Polymerase chain reaction
Pekin et al49 developed a droplet-based microfluidic system based on digital PCR to perform highly sensitive and quantitative detection of mutations in the KRAS oncogene, a tumor biomarker. The device was fabricated with PDMS, and a hydrophobic coating was applied to the inside wall of the microchannels to facilitate droplet generation. The genomic DNA was encapsulated in 9 pL droplets along with two fluorogenic TaqMan® probes, which generated green fluorescent droplets for mutant DNA and red ones for the wild-type DNA. This system enabled the detection of a KRAS mutated gene in a large excess (200,000×) amount of wild-type KRAS genes from cultured cells. By adding probes corresponding to different mutations, this system allowed the simultaneous screening of six common mutations in the KRAS codon in one single run. To test the clinical utility, this system was used to screen seven common KRAS mutations using blood plasma samples from patients with metastatic colorectal cancer (CRC) and successfully identified 14 of the 19 confirmed KRAS mutation cases. This showed the promise for a noninvasive diagnostic technique using only blood plasma instead of biopsies.50 Klein et al51 presented a high-throughput microfluidic platform for droplet barcoding and analysis of stem cells before and after leukemia inhibitory factor withdrawal. The microfluidic device was fabricated with PDMS by soft lithography, and the depth of microchannels was 80 µm (Figure 2A). Mouse embryonic cells were encapsulated in droplets with the lysis buffer and reverse transcription (RT) reagents along with hydrogel microspheres carrying barcoding ssDNA. After the cell lysis and RT reaction, the barcoded complementary DNA was sequenced to study the cell population structure and heterogeneous onset of differentiation under leukemia inhibitory factor influence. This platform allowed randomly labeling up to 3,000 cells with 99% unique labels for every 100 µL emulsion (12,000 cells/h) and could be used to identify cell types based on gene expression for clinical samples, such as tumors and tissue microbiopsies. Ferraro et al52 recently developed a new fully automated, programmable microfluidic platform by integrating droplet RT-quantitative PCR and magnetic tweezers to determine the expression level of the human epidermal growth factor receptor 2 (HER2), a biomarker for the breast cancer, by the quantification of the messenger RNA. The microfluidic chips were fabricated with PDMS and connected to two syringe pumps for droplet generation. The RNA purification and RT-PCR were performed in droplets of 250 nL. This platform was tested with samples from both cultured cells and clinically validated patients with breast cancers, and the results were in reasonable agreement with those from conventional tests. Compared to the currently used motorized platforms, the sample consumption for this system was decreased by 100 times, and it only took 3 min to process a sample, which allowed high-throughput applications.
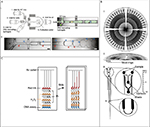  | Figure 2 Microfluidic devices for nucleic acid detection. Notes: (A) A microfluidic device for droplet barcoding ribonucleic acid from individual cells for sequencing. (i) Design of the microfluidic device with microchannels of 80 µm depth (left) and scheme of the on-chip encapsulation process (right). (ii) Micrographs of encapsulation (left) and collection (right) modules. Cells, hydrogel microspheres, and flow direction are indicated by red, blue, and black arrows, respectively. The scale bars represent 100 µm. (B) Schematic views of the microfluidic polymorphism ratio sequencing plate. (i) The top view of the plate containing 96 separation capillaries with a common anode at the center. (ii) Cross-sectional view of the plate. A final layer of 1/8 in. thick Borofloat rings was attached to the top of the plate as the moat rings. (iii, iv) Expanded view of an individual capillary. Each capillary was 17 cm long, 200 µm wide, and 30 µm deep. (C) Schematic view of the multistage volumetric bar chart chip for multiplex, quantitative DNA detection. The oxygen generated during the DNA assay pushes the red ink in the microchannels for visual readouts. (A) Reprinted from Cell, 161(5), Klein AM, Mazutis L, Akartuna I, et al, Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells, 1187–1201, Copyright 2015, with permission from Elsevier.51 (B) Adapted from Paegel BM, Emrich CA, Wedemayer GJ, Scherer JR, Mathies RA. High throughput DNA sequencing with a microfabricated 96-lane capillary array electrophoresis bioprocessor. Proc Natl Acad Sci. 2002;99(2):574–579. Copyright © 2002, The National Academy of Sciences.56 (C) Adapted with permission from Song Y, Wang Y, Qin L. A multistage volumetric bar chart chip for visualized quantification of DNA. J Am Chem Soc. 2013;135(45):16785–16788. Copyright © 2013 American Chemical Society.58 Abbreviation: RT, reverse transcription. |
Gel electrophoresis
Zhang et al53 developed a microchip-based temperature gradient CE system for the detection of low-abundance K-ras mutation, a biomarker for the epidermal growth factor receptor-targeted therapy in CRCs. The microfluidic chip was fabricated with glass and had a cross-channel design with sample loading and separation channels of 1.0 and 4.5 cm, respectively. This system was tested with clinical stool samples from CRC patients. Among the 30 stool samples, 17 (57%) were identified to contain K-ras mutations and the detection limit in the stool samples was 2%. The coincidence rate for K-ras mutations between tissues and stools was 97%. This system featured short analysis time (6 min), low sample consumption (14 nL), and high sensitivity (0.2% mutant CRC cells in a wild-type background). It showed promising results for predicting the effectiveness of epidermal growth factor receptor-targeted therapy for CRCs by noninvasive means. Duberow et al54 described a microfluidic polymorphism ratio sequencing (PRS) system to detect somatic D-loop mutations in the mitochondrion DNA, a biomarker for urothelial cell carcinoma (UCC). PRS is based on the Sanger sequencing and allows direct and quantitative determination of sequence variations.55 The microfluidic device was fabricated with a 150 mm Borofloat glass wafer and contained 96 separation capillaries converging on a common anode (Figure 2B).56 Genomic DNA was extracted from 14 clinically validated UCC tissue specimens and further processed for PRS extension and pooling. The PRS fragments were then separated by microchip electrophoresis for 30 min. The authors analyzed the D-loop mitochondrion DNA regions in 14 primary UCC tumors to identify a total of 28 somatic mutations. Among the 14 samples, 8 (57%) were identified to contain at least one somatic D-loop mutation. This system enabled high-throughput mutation screening through parallelized analysis with very low sample consumption and short analysis times.
Hybridization
Ferguson et al57 presented a magnetic integrated microfluidic electrochemical detector (MIMED) for the detection of H1N1 influenza viruses directly from throat swab samples. The MIMED device (1×6 cm) was fabricated with glass and PDMS and had a sample preparation chamber (35 µL) and an electrochemical DNA detection cell (7 µL). The target capture, concentration and purification, RT-PCR amplification, and ssDNA generation were performed in the sample preparation chamber. The virus detection was achieved by monitoring variations in the faradic current caused by the sequence-specific conformation changes during the target DNA hybridization. The whole process took ~3.5 h to complete from sample to answer, with the RT-PCR being the limiting step (~150 min). The limit of detection (LOD) of this system was ~10 nM ssDNA from throat swab samples, which was 4 orders of magnitude below clinical viral titers and >2 orders of magnitude lower than rapid tests for swine influenza viruses. Song et al58 developed a multistage volumetric bar chart chip (MV-Chip) for multiplex quantitative DNA detection. The microfluidic chip was fabricated using two glass slides (75×50×1.0 mm). It contained 50 µm deep microchannels for parallel assays (six or ten targets at a time) and on-chip platinum microelectrodes (Figure 2C). The assay was carried out by DNA hybridization, which introduced an enzyme probe to initiate the production of oxygen that pushed the red ink in microchannels for visual readouts. Longer red ink lines indicated higher DNA concentrations. The LOD was 20 pM DNA. The test results could be read by naked eyes without the need for complicated instruments. Heo et al59 designed and constructed a valveless rotary microfluidic chip for multiplex detection of the TP53 gene mutations, which were reported to take place in many cancers. The microfluidic chip was composed of three glass layers: a rotary wafer with 12 reaction chambers, a microchannel wafer, and a temperature detector wafer with Ti/Pt electrodes. The authors designed five DNA padlock probes to detect point mutations corresponding to cancer prognosis in the single nucleotide polymorphism sites of the TP53 gene. By turning the rotary wafer to different positions, a series of sample injections, ligation-rolling circle amplification reactions, and fluorescence detection of the amplicons were performed to identify the target mutation points. The whole assay took 120 min to complete, and the LOD was 1 fmol DNA. This microfluidic chip allowed multiplex single nucleotide polymorphism analysis without complicated fluid control involving valves, pumps, and tubing lines.
Microfluidic devices for protein-based tests
Proteins can serve as valuable biomarkers because of their high specificity, involvement in many biological processes, and capability to carry more information after alternative splicing and posttranslational modifications. In addition, many physiologic changes, for example, signal transduction, cell differentiation, and malignant transformation, are controlled posttranscriptionally and do not show at the nucleic acid level. As a result, proteins can provide important information on the biological states by their level of abundance and structural/functional changes in a biological system.60 However, there are challenges for microfluidics-based protein detection. First, proteins are much more diverse than DNAs and RNAs in both numbers (~300,000 to several millions) and sizes (tens to ~36,000 amino acids).61,62 Second, proteins of clinical interest are usually in low abundance compared to those on the background in biological samples, while there is no amplification process equivalent to PCR for nucleic acids. To address these challenges, immunoaffinity-based techniques (“immunoassays”) have been miniaturized onto microfluidic devices for detection and quantification of target proteins by exploiting the high sensitivity and specificity of antibody–antigen interactions.63 As described in the “Microfluidic devices for nucleic acid-based tests” section, gel electrophoresis is also applicable to the separation and detection of proteins for diagnostic applications.
Immunoassays
Chikkaveeraiah et al64 developed a microfluidic electrochemical immunoarray for two cancer biomarkers, prostate-specific antigen and interleukin-6 (IL-6), in human serum samples. The chip was composed of a PDMS microchannel sealed onto a disposable eight-electrode microarray with immobilized capture antibodies. The cancer biomarkers were first extracted from serum samples with magnetic microbeads functionalized with enzyme labels and antibodies. The complex was then loaded into the microchannel to be captured onto the electrode surfaces. The detection and quantification of the biomarkers were achieved by amperometric signals resulting from the electron transfer between electrodes and enzyme labels, which was activated by injecting a solution of mediator and hydrogen peroxide. This assay required only 5 µL of patient serum and 69 min to complete. The LOD was 0.23 pg/mL for prostate-specific antigen and 0.30 pg/mL for IL-6. Later, the same group further optimized its microfluidic array for the simultaneous detection and quantification of four cancer biomarkers: IL-6, IL-8, vascular endothelial growth factor (VEGF), and VEGF-C in diluted serum. The assay time was reduced to 50 min. The LOD reached a range of 5–50 fg/mL.65
In clinical applications, enzyme-linked immunosorbent assay (ELISA) is broadly used as a diagnostic tool for its precision, sensitivity, versatility, and quantifiability. Conventional ELISA protocols require multiple steps of adsorption, washing, and incubation of reagents.66 Chin et al67 developed a microfluidic ELISA chip, the “mChip”, for the detection of human immunodeficiency virus and syphilis in resource-limited settings. The microfluidic chips were fabricated with transparent polystyrene and cyclic olefin copolymer by injection molding (Figure 3A). The steps of reagent addition and washing were carried out by a bubble-based delivery, where measured plugs of reagents and washing buffers were loaded sequentially onto the chip in a tubing with air bubbles as spacers. A manual syringe was used to provide the vacuum needed for fluid manipulations. It took only 1 µL of unprocessed whole blood and 20 min to complete the diagnosis of human immunodeficiency virus. Signal detection was done with a compact device consisting of low-cost light-emitting diodes (LEDs; $0.50 USD each) and photodetectors ($6.00 USD each). Compared to lab-based ELISA, the mChip had a much lower requirement for equipment and could be easily deployed in remote areas for rapid screening. Park et al68 reported a fully automatic lab-on-a-disk system for the simultaneous detection of cardiovascular disease biomarkers (high-sensitivity C-reactive protein, cardiac troponin I, and N-terminal pro-B type natriuretic peptide) from whole blood and whole saliva. This system included the lab-on-a-disk and a detection module composed of an LED and a photodiode for absorbance measurements. The disk was made with cyclic olefin copolymer for optical clarity. Microchannels and reaction chambers were fabricated on the bottom layer by CNC micromachining, while the top layer had access ports and ferrowax loading chambers. Before assembling with the double-sided tape, polystyrene beads functionalized with capture antibodies were loaded into the main reaction chambers for a bead-based sandwich ELISA. Fluid manipulations were carried out by the centrifugal force and reversible phase changes of the ferrowax valves. Each disk could be used for the fully automated analysis of six biomarkers from two samples in 20 min and only required a total sample volume of 200 µL. For saliva samples, the LOD was 0.30, 0.51, and 0.24 ng/mL for high-sensitivity C-reactive protein, cardiac troponin I, and N-terminal pro-B type natriuretic peptide, respectively. For blood samples, the LOD was 0.27, 0.27, and 0.32 ng/mL, respectively. This system could be revised for other immunoassays by simply changing the capture antibodies. Li et al69 developed a nanoporous glass-integrated volumetric bar-chart chip to detect the three biomarkers (CEA, CYFRA 21-1, and SCCA) for non-small cell lung cancer. The chip was composed of a nanoporous glass membrane sandwiched between two glass slides with microstructures fabricated using standard photolithographic techniques (Figure 3B). The membrane had multiple nanopores (average diameter=200 nm) that significantly increased the surface area for immobilizing capture antibodies for the sandwich ELISA reaction. Each chip allowed four assays to run simultaneously. The detection and quantification of the target protein biomarkers were visually represented by the travel distance of red ink bars, which were propelled by the oxygen generated from the reaction between the ELISA probe and H2O2. The nanoporous glass-integrated volumetric bar-chart chip was used to test 21 serum samples from cancer patients, and the results were consistent with those acquired from commercial ELISA kits. The LOD was 50 pg/mL, and the total analysis time was 30 min.
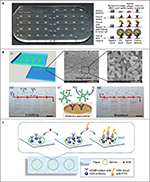  | Figure 3 Microfluidic devices for protein detection. Notes: (A) Microfluidic ELISA chip and the schematic illustration of HIV and syphilis detection. Each chip can process seven samples simultaneously. (B) Nanoporous glass-integrated volumetric chip for cancer biomarker detection. (i–iii) The chip design and scanning electron microscope images of the nanoporous glass membrane with scale bars representing 5 μm (ii) and 1 μm (iii). (iv–vi) Sample loading, sandwich ELISA, and result readout. Scale bars=1 cm for (iv) and (vi), respectively. (C) Paper-based analytical device for CEA detection based on FRET using doped UCNPs. (A) Adapted by permission from Macmillan Publishers Ltd: Nat Med. Chin CD, Laksanasopin T, Cheung YK, et al. Microfluidics-based diagnostics of infectious diseases in the developing world. 2011;17(8):1015–1019. Copyright © 2011, Rights Managed by Nature Publishing Group. http://www.nature.com/nm/index.html.67 (B) Adapted with permission from Li Y, Xuan J, Song Y, et al. Nanoporous glass integrated in volumetric bar-chart chip for point-of-care diagnostics of non-small cell lung cancer. ACS Nano. 2016;10(1):1640–1647. Copyright © 2016 American Chemical Society.69 (C) Adapted from Xu S, Dong B, Zhou D, et al. Paper-based upconversion fluorescence resonance energy transfer biosensor for sensitive detection of multiple cancer biomarkers. Sci Rep. 2016;6:23406. Copyright © 2016, Rights Managed by Nature Publishing Group. The Creative Commons license is available at: https://creativecommons.org/licenses/by/4.0/legalcode.74 Abbreviations: BSA, bovine serum albumin; CEA, carcinoembryonic antigen; ELISA, enzyme-linked immunosorbent assay; FITC, fluorescein isothiocyanate; FRET, fluorescence energy transfer; IgG, immunoglobulin G; PtNP, platinum nanoparticle; UCNPs, upconversion nanoparticles. |
First introduced by the Whitesides group,70,71 paper (cellulose or cellulose–polymer blends) has been used to fabricate microfluidic devices for bioassays because of its high abundance, low cost, biocompatibility, ease of use/storage, and fluid transport by capillary action without active pumping. Wang et al72 developed a wax-patterned, three-dimensional microfluidic paper-based electrochemical device for the detection of tumor biomarkers, carcinoma antigen 125 (CA125) and carcinoembryonic antigen (CEA), in real human serum samples by electrochemical immunoassays. The three-dimensional microfluidic paper-based electrochemical device consisted of a top wax-patterned layer as an insulator and a bottom substrate layer with printed electrodes. The detection and quantification of the biomarkers were achieved by amperometric signals generated by a sandwich immunoreaction. The LOD was 0.2 mU/mL and 0.01 ng/mL for CA125 and CEA, respectively. Wu et al73 developed a microfluidic paper-based electrochemical immunodevice with integrated graphene film for signal amplification to detect and quantify four cancer biomarkers, alpha-fetoprotein, CEA, CA125, and carbohydrate antigen 153, in serum samples from cancer patients. The immunodevice consisted of two layers of chromatography paper selectively patterned with the SU-8 3010 photoresist. Working electrodes were screen-printed onto the two layers for signal transmission. The LOD was 0.001, 0.005, 0.001, and 0.005 ng/mL for alpha-fetoprotein, CEA, CA125, and carbohydrate antigen 153, respectively. After regeneration, the immunodevice could be reused for at least 24 times without any obvious loss in performance. Xu et al74 presented a paper-based biosensor for the detection of CEA, a cancer biomarker. The biosensor was fabricated by patterning on a piece of filter paper with CuS nanoparticle (upconversion nanoparticles) solution using an HP commercial printer (Figure 3C). The detection and quantification of the biomarker were done through the response of the fluorescence resonance energy transfer on the device. The LOD was 0.89 ng/mL. One possible disadvantage of paper-based microfluidic devices is that they need careful handling for stability and reproducibility, because paper and patterns on it (such as electrodes) are fragile. A device can be damaged by bending, folding, tearing, scratching, or long-term soaking in sample/reagent solutions. More in-depth information on paper-based microfluidic devices and their applications can be found in recent reviews and the references cited therein.75–77
Gel electrophoresis
Huang et al78 developed a method based on microchip electrophoresis coupled with laser-induced fluorescence detection to phenotype haptoglobin (Hp), a biomarker for the liver cancer. The microfluidic device was fabricated with glass. The microchannels were 25 µm wide and 50 µm deep and had an effective separation length of 4 cm. Hp was labeled with the fluorescein isothiocyanate dye (excitation wavelength=488 nm, emission wavelength=520 nm) for laser-induced fluorescence detection. The process of separation and detection took 150 s to complete, and the LOD was 0.39 and 0.62 µg/mL for two Hp phenotypes, Hp1-1 and 2-2, respectively. Serum samples from healthy individuals and liver cancer patients were tested, and a decrease in the Hp concentration was confirmed for liver cancer patients. This study demonstrated the potential of a simple, efficient, and fast method for disease diagnosis and proteome research. Kalish and Phillips79 used a microchip-based immunoaffinity CE system to detect the presence of CXC chemokines, biomarkers for atopic dermatitis, in frozen biopsy samples from patients. Target CXC chemokines were captured with a disposable immunoaffinity disk, labeled with a fluorescence dye, and separated by microchip electrophoresis. The microfluidic chip was fabricated with glass and had 50 µm wide, 20 µm deep microchannels. The separation length was 35 mm. The whole assay took ~40 min, including the biopsy microdissection, sample pretreatment, and microchip electrophoresis. The LOD ranged from 0.2 to 0.4 pg/ mL for each of the six chemokines tested in this study. This system enabled the detection and quantification of chemokines relevant to inflammatory skin lesions in clinical histopathology samples and featured very low sample consumption (200 nL), decreased analysis time, and higher sensitivity. Lin et al80 developed a method for multiplex protein assay based on tunable aptamer and microchip electrophoresis. Aptamers are ssDNA or single-stranded RNA molecules that can specifically bind to preselected targets such as proteins, cells, and metal ions. In this study, they were used to form aptamer–protein complexes that could be separated by microchip electrophoresis according to mobility differences. The microfluidic device was fabricated with quartz and had 88 µm wide, 50 µm deep microchannels with an effective separation length of 23 mm. Three cancer-relevant proteins, thrombin, platelet-derived growth factor-BB (PDGF-BB), and vascular endothelial growth factor 165 (VEGF165), were successfully separated and quantified on this microfluidics-based system. The whole process took 185 s to complete and only required a very low sample volume (2 µL). The LOD was 4.1 nM for thrombin, 1.96 nM for PDGF-BB, and 2.48 nM for VEGF165. The dynamic range was between 5 and 150 nM. This method allowed multiplex protein analysis and could be extended to other biological molecules that can bind to aptamers. In addition to nucleic acids and proteins, lipids, carbohydrates, and metabolites can also serve as useful biomarkers. More in-depth information can be found in several recent reviews and the references cited therein.81–83
Transitioning to point-of-care testing
As supported by the work reviewed so far, molecular diagnostic tests can be successfully miniaturized onto microfluidic devices and demonstrated enhanced performance compared to their counterparts at the macroscale. However, there are additional requirements to be considered for building viable diagnostic instruments based on these microfluidic devices for routine clinical applications in the field. Conventional diagnostic instrumentation generally requires established infrastructure, reliable electricity supply, costly reagents, long run time, and trained professionals to operate, which are usually unavailable in developing countries and resource-limited environments. To effectively deliver health care, particularly for infectious disease control, there is a strong demand for the capability of performing rapid and timely diagnostic tests at or near the site of patient care (point-of-care testing [POCT]).84 Developed countries with established health care systems can also benefit from POCT through improved preventive and personalized health care made possible by easily and frequently performing diagnostic assays. For example, the detection and quantification of lipoprotein biomarkers can help better predict the risk of cardiovascular diseases, and the use of the information on specific tumor biomarkers can help doctors develop personalized treatment plans for cancers based on individual needs.85,86 To be viable, the platform for POCT must be portable, inexpensive, and easy to operate by nonspecialists without the need for complex equipment and established infrastructure. The assays used must be accurate, reliable, reproducible, and fast in terms of seconds and minutes. The benefits of microfluidics, such as portability, reduction in fabrication costs and sample/reagent volumes, and significant increase in sample process speed, are a natural fit for developing POCT devices. Although there are many successful examples of bioassays miniaturized onto microfluidic devices, many of these microchips still require trained professionals to operate and connections to external equipment, such as microscopes, computers, and fluid pumping systems, to perform their tasks, which make it a major roadblock to further deploy POCT systems for better health care. The solution to this roadblock comes from the mass production of open-source microcontrollers (e.g., Raspberry Pi and Arduino) and consumer electronics devices (e.g., tablets and smartphones). Smartphones are an ideal platform for developing POCT systems, because they mostly come with digital cameras, various sensors, and graphics processing and computing units, along with their coverage and data connectivity almost everywhere in the world (~97% global penetration rate).87 Here, we review selected microfluidics-based POCT systems developed by the integration of miniaturized molecular techniques, open-source electronics, and smartphones.
Wang et al88 developed a microchip ELISA-based system with cell phone imaging to quantify an ovarian cancer biomarker, HE4, in clinical urine samples. The microfluidic device was fabricated with poly(methyl methacrylate) and double-sided adhesive films. Each straight microchannel had access ports on both ends and dimensions of 4×7.5×3.225 mm (Figure 4A). Microchip ELISA was carried out by manually loading samples and reagents into the microchannel for incubation. A cell phone with a customized mobile application was used to acquire the color intensity of the ELISA solution in the microchannel after incubation and to report the results on the screen after data processing. The whole assay required 100 µL of urine samples and 15 min to complete with a detection limit of 19.5 ng/mL. Mancuso et al89 developed a microfluidics-based smartphone accessory for the detection of Kaposi’s sarcoma associated herpesvirus (KSHV) nucleic acids. The microfluidic device was with PDMS and glass and had a 1 cm long microchannel for absorbance measurements. The phone accessory for absorbance measurements consisted of a magnetic latch, LED, photodetector, and an Arduino board that controlled the LED and photodetector (Figure 4B). The phone accessory obtained power from and communicated with the phone through a male micro-USB connector. A customized mobile application was coded to control the whole system and to display results on the screen. KSHV probes were prepared by conjugating gold nanoparticles (average diameter=15 nm) with oligonucleotides specific to KSHV DNA. The assay was carried out by mixing the probe solution with samples containing the KSHV DNA. After 24 h of incubation, the mixture was loaded onto the microchip for absorbance measurement. The LOD was 1 nM of KSHV DNA. Instead of the common handheld devices, Priye et al90 constructed a lab-on-a-drone system that could perform in-flight PCR isothermally for nucleic acid-based diagnosis (Figure 4C). The analysis instrument consisted of a hand-crank power source, an isothermal heater, a blue excitation LED, a liquid crystal display screen, an Arduino board, and a PCR reactor made with a polycarbonate rod (~20 µL in volume). A smartphone with a customized mobile application was used to perform the fluorescence detection of Staphylococcus aureus and λ-phage DNA targets. The whole assay took 20 min to complete and had a detection limit of 1,000 copies/µL. The team also designed and 3D-printed rotor attachments that could be connected to the drone motors to perform centrifuge-based sample processing. This system made it possible to rapidly deploy nucleic acid-based diagnostics using consumer quadcopter drones at a low cost. More in-depth information on POCT systems can be found in recent reviews and the references cited therein.91–93 As supported by the success from academic research, there is great potential for microfluidic molecular diagnostics to be commercially viable.
  | Figure 4 Point-of-care testing systems. Notes: (A) Detection of the ovarian cancer biomarker, HE4, by integrating microchip ELISA with cell phone-based colorimetric analysis. (i) Sample and reagent loading. (ii) On-chip sandwich ELISA for color development. (iii) Image acquisition with the built-in camera. (iv) Data processing and result report on the cell phone. (B) Configuration of the smartphone accessory for the detection of Kaposi’s sarcoma associated herpesvirus nucleic acids. (i) An image of the assembled smartphone-based system. (ii) The internal layout of the smartphone accessory. (iii) A sample microfluidic cartridge with 1 cm optical path for sample detection. (C) Configuration of lab-on-a-drone. (A) Adapted from Wang S, Zhao X, Khimji I, et al. Integration of cell phone imaging with microchip ELISA to detect ovarian cancer HE4 biomarker in urine at the point-of-care. Lab Chip. 2011;11(20):3411–3418. With permission from The Royal Society of Chemistry. © The Royal Society of Chemistry 2011. http://dx.doi.org/10.1039/C1LC20479C.88 (B) Adapted from Mancuso M, Cesarman E, Erickson D. Detection of Kaposi’s sarcoma associated herpesvirus nucleic acids using a smartphone accessory. Lab Chip. 2014;14(19):3809–3816. With permission from The Royal Society of Chemistry. © The Royal Society of Chemistry 2014. http://dx.doi.org/10.1039/C4LC00517A.89 (C) Adapted with permission from Priye A, Wong S, Bi Y, et al. Lab-on-a-drone: toward pinpoint deployment of smartphone-enabled nucleic acid-based diagnostics for mobile health care. Anal Chem. 2016;88(9):4651–4660. Copyright © 2016 American Chemical Society. http://pubs.acs.org/doi/full/10.1021/acs.analchem.5b04153. Permission requests for Figure C should be directed to the American Chemical Society.90 Abbreviations: BSA, bovine serum albumin; ELISA, enzyme-linked immunosorbent assay; HE4, human epididymis protein 4; HRP, horseradish peroxidase; LED, light-emitting diode; PCR, polymerase chain reaction; TMB, tetramethylbenzidine. |
Challenges to commercialization
Commercialization plays a critical role in making microfluidic molecular diagnostics accessible to both health care professionals and regular end users. In 2015, the global molecular diagnostics market was valued at $6.45 billion USD and is projected to reach 17.9 billion USD by 2024 with an expected compound annual growth rate of 12%,94 representing a very large opportunity for microfluidic molecular diagnostics. With advances in microfluidics technology and chemical/biological techniques over the past two decades, a number of companies have made great efforts to translate results from academic research into commercial products (Table 1).95,96 However, there are relatively few commercial products available in the market, given the immense interest from both academia and industry. This is because of the lack of a game-changing product (or a “killer application”) that can generate at least hundreds of millions of dollars in revenue,97 and thus, many investors would turn to other emerging technologies potentially coming with high revenues, resulting in reduced financial support for new product development. In addition, the fabrication costs remain high, when a product does not have high volume demand to reach economies of scale.
  | Table 1 List of selected products for microfluidics-based, molecular diagnostic products in the market Abbreviation: PCR, polymerase chain reaction. |
To overcome the barriers to commercialization, researchers need to address two major challenges. 1) Component integration: previous research efforts have been focused on miniaturizing molecular diagnostic techniques onto microfluidic devices, while very little consideration has been given to how they are integrated with other components, such as sample pretreatment, fluid control, power supply, and data acquisition/processing, to make a self-contained, functional instrument. Without careful planning for integration, any efforts to optimize individual components can make them mutually incompatible, when putting them together in the final product. To address this, researchers should consider the final commercial application in the beginning of instrument design and ensure that all components are compatible for integration. 2) Fabrication standardization: for academic research, there are no standard materials and processes for developing microfluidic devices for molecular diagnostic tests. The choice of device materials and fabrication processes usually depends on the target application, which makes it difficult for commercialization, because popular materials in academia are often not compatible with mass production. For example, PDMS is a very popular material for microfluidic devices. However, it is very costly when compared to the common polymers used for industrial applications, such as polycarbonate and poly(methyl methacrylate), and soft lithography is difficult to be fully automated. To address this, researchers need to work with manufacturers to identify suitable materials and fabrication processes, when developing products for microfluidic molecular diagnostics.
Summary and outlook
For over two decades, a lot of progress has been made in the field of microfluidics, ranging from the science of studying and controlling the behaviors of fluids and particles at microscale to the technology of developing methods and devices that can precisely and effectively perform such tasks. For applications in molecular diagnostics, many bioassays have been successfully miniaturized onto microfluidic devices, with performance better than that of their benchtop counterparts. The commercialization of these bioassays has been moving forward faster over the past few years, driven by the need for full-scale integration and automation of all microfluidic components onto a single instrument that can run a process from sample preparation to final results, that is, the “sample in, answer out” platform. More collaborative and interdisciplinary efforts between academia and industry are still needed to develop true “all-in-one” microfluidic devices that can carry out molecular diagnosis in a simple, cost-effective way. In addition to technical advances, it is also critical to identify suitable markets for microfluidic molecular diagnostics, particularly niche markets where conventional technologies cannot meet the special requirements from potential customers. The industrial parties with marketing skills should be active in surveying end-user requirements and work with their academic partners based on the feedback to develop products that best address these special requirements. This would significantly help microfluidic molecular diagnostics reach its maximum potential for both commercialization and social impact.
In summary, microfluidics technology, as an enabling platform, has a promising outlook for diagnostic applications because of its potential to make laboratory-based diagnostics more accessible to concerned parties for streamlined health care, improved clinical outcomes, and rapid intervention. We can expect that future advances, particularly in POCT, will have a transformative impact on the global health care system to improve our quality of life, especially for people in developing countries and resource-limited environments.
Acknowledgments
The author thanks the Department of Chemical Engineering and College of Engineering at California State University, Long Beach (CSULB) for their support. This work was supported in part by the CSULB Research, Scholarship, and Creativity Awards (RSCA) and the CSULB Sabbatical Leave Award.
Disclosure
The author reports no conflicts of interest in this work.
References
Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442(7101):368–373. | ||
Burns MA, Johnson BN, Brahmasandra SN, et al. An integrated nanoliter DNA analysis device. Science. 1998;282(5388):484–487. | ||
Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298(5593):580–584. | ||
West J, Becker M, Tombrink S, Manz A. Micro total analysis systems: latest achievements. Anal Chem. 2008;80(12):4403–4419. | ||
Erickson D, Li D. Integrated microfluidic devices. Anal Chim Acta. 2004;507(1):11–26. | ||
Golay MJE. Vapor phase chromatography and telegrapher’s equation. Anal Chem. 1957;29(6):928–932. | ||
van Deemter JJ, Zuiderweg FJ, Klinkenberg A. Longitudinal diffusion and resistance to mass transfer as causes of nonideality in chromatography. Chem Eng Sci. 1995;50(24):3869–3882. | ||
Luckey JA, Drossman H, Kostichka AJ, et al. High speed DNA sequencing by capillary electrophoresis. Nucleic Acids Res. 1990;18(15):4417–4421. | ||
Petersen KE. Silicon as a mechanical material. Proc IEEE. 1982;70(5):420–457. | ||
Terry SC, Jerman JH, Angell JB. A gas chromatographic air analyzer fabricated on a silicon wafer. IEEE Trans Electron Devices. 1979;26(12):1880–1886. | ||
de Mello AJ. FOCUS On-chip chromatography: the last twenty years. Lab Chip. 2002;2(3):48N–54N. | ||
Manz A, Miyahara Y, Miura J, Watanabe Y, Miyagi H, Sato K. Design of an open-tubular column liquid chromatograph using silicon chip technology. Sens Actuators B Chem. 1990;1(1–6):249–255. | ||
Reston RR, Kolesar ES. Silicon-micromachined gas chromatography system used to separate and detect ammonia and nitrogen dioxide. I. Design, fabrication, and integration of the gas chromatography system. J Microelectromechanical Syst. 1994;3(4):134–146. | ||
Kolesar ES, Reston RR. Silicon-micromachined gas chromatography system used to separate and detect ammonia and nitrogen dioxide. II. Evaluation, analysis, and theoretical modeling of the gas chromatography system. J Microelectromechanical Syst. 1994;3(4):147–154. | ||
Lin S-L, Lin T-Y, Fuh M-R. Microfluidic chip-based liquid chromatography coupled to mass spectrometry for determination of small molecules in bioanalytical applications: an update. Electrophoresis. 2014;35(9):1275–1284. | ||
Haghighi F, Talebpour Z, Sanati-Nezhad A. Through the years with on-a-chip gas chromatography: a review. Lab Chip. 2015;15(12):2559–2575. | ||
Manz A, Graber N, Widmer HM. Miniaturized total chemical analysis systems: a novel concept for chemical sensing. Sens Actuators B Chem. 1990;1(1–6):244–248. | ||
Manz A, Harrison DJ, Verpoorte EMJ, Fettinger JC, Lüdi H, Widmer HM. Miniaturization of chemical analysis systems a look into next century’s technology or just a fashionable craze? Chim Int J Chem. 1991;45(4):103–105. | ||
Manz A, Harrison DJ, Verpoorte EMJ, et al. Planar chips technology for miniaturization and integration of separation techniques into monitoring systems: capillary electrophoresis on a chip. J Chromatogr A. 1992;593(1–2):253–258. | ||
Vasudev A, Kaushik A, Jones K, Bhansali S. Prospects of low temperature co-fired ceramic (LTCC) based microfluidic systems for point-of-care biosensing and environmental sensing. Microfluid Nanofluidics. 2012;14(3–4):683–702. | ||
Yu X, Tu S-T, Wang Z, Qi Y. Development of a microchannel reactor concerning steam reforming of methanol. Chem Eng J. 2006;116(2):123–132. | ||
Ren K, Dai W, Zhou J, Su J, Wu H. Whole-teflon microfluidic chips. Proc Natl Acad Sci U S A. 2011;108(20):8162–8166. | ||
Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew Chem Int Ed. 2007;46(8):1318–1320. | ||
Xia Y, Whitesides GM. Soft Lithography. Annu Rev Mater Sci. 1998;28(1):153–184. | ||
Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3(1):335–373. | ||
Friend J, Yeo L. Fabrication of microfluidic devices using polydimethylsiloxane. Biomicrofluidics. 2010;4(2):026502-1–026502-5. | ||
Chin CD, Laksanasopin T, Cheung YK, et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat Med. 2011;17(8):1015–1019. | ||
Focke M, Kosse D, Müller C, Reinecke H, Zengerle R, Stetten F von. Lab-on-a-foil: microfluidics on thin and flexible films. Lab Chip. 2010;10(11):1365–1386. | ||
Cameron NS, Roberge H, Veres T, Jakeway SC, Crabtree HJ. High fidelity, high yield production of microfluidic devices by hot embossing lithography: rheology and stiction. Lab Chip. 2006;6(7):936–941. | ||
Mair DA, Geiger E, Pisano AP, Fréchet JMJ, Svec F. Injection molded microfluidic chips featuring integrated interconnects. Lab Chip. 2006;6(10):1346–1354. | ||
Hull CW, Uvp I. Apparatus for production of three-dimensional objects by stereolithography; 1984. Available from: https://www.google.com/patents/US4575330. Accessed June 2, 2016. | ||
Crump SS, Stratasys I. Apparatus and method for creating three-dimensional objects; 1989. Available from: https://www.google.com/patents/US5121329. Accessed June 2, 2016. | ||
Waheed S, Cabot JM, Macdonald NP, et al. 3D printed microfluidic devices: enablers and barriers. Lab Chip. 2016;16(11):1993–2013. | ||
Au AK, Huynh W, Horowitz LF, Folch A. 3D-Printed Microfluidics. Angew Chem Int Ed. 2016;55(12):3862–3881. | ||
Ho CMB, Ng SH, Li KHH, Yoon Y-J. 3D printed microfluidics for biological applications. Lab Chip. 2015;15(18):3627–3637. | ||
Yazdi AA, Popma A, Wong W, Nguyen T, Pan Y, Xu J. 3D printing: an emerging tool for novel microfluidics and lab-on-a-chip applications. Microfluid Nanofluidics. 2016;20(3):1–18. | ||
Bhattacharjee N, Urrios A, Kang S, Folch A. The upcoming 3D-printing revolution in microfluidics. Lab Chip. 2016;16(10):1720–1742. | ||
Gomez FA, ed. Biological Applications of Microfluidics. 1st ed. Hoboken, NJ: Wiley-Interscience; 2008. | ||
Tian W-C, Finehout E. Microfluidics for Biological Applications.1st ed. Springer: US; 2009. Available from: http://www.springer.com/us/book/9780387094793. Accessed June 2, 2016. | ||
Yeo LY, Chang H-C, Chan PPY, Friend JR. Microfluidic devices for bioapplications. Small Weinh Bergstr Ger. 2011;7(1):12–48. | ||
Sackmann EK, Fulton AL, Beebe DJ. The present and future role of microfluidics in biomedical research. Nature. 2014;507(7491):181–189. | ||
Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS. 2010;5(6):463. | ||
Leitner E, Kessler HH, Spindelboeck W, et al. Comparison of two molecular assays with conventional blood culture for diagnosis of sepsis. J Microbiol Methods. 2013;92(3):253–255. | ||
Mancini N, Carletti S, Ghidoli N, Cichero P, Burioni R, Clementi M. The era of molecular and other non-culture-based methods in diagnosis of sepsis. Clin Microbiol Rev. 2010;23(1):235. | ||
Grody WW, Nakamura RM, Kiechle FL, Strom C, eds. Molecular Diagnostics: Techniques and Applications for the Clinical Laboratory. 1st ed. Academic Press: London; 2009. | ||
Netto GJ, Saad RD, Dysert PA, II. Diagnostic molecular pathology: current techniques and clinical applications, part I. Proc Bayl Univ Med Cent. 2003;16(4):379. | ||
Landers JP. Molecular diagnostics on electrophoretic microchips. Anal Chem. 2003;75(12):2919–2927. | ||
Ugaz VM, Elms RD, Lo RC, Shaikh FA, Burns MA. Microfabricated electrophoresis systems for DNA sequencing and genotyping applications: current technology and future directions. Philos Trans R Soc Lond Math Phys Eng Sci. 2004;362(1818):1105–1129. | ||
Pekin D, Skhiri Y, Baret J-C, et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip. 2011;11(13):2156–2166. | ||
Taly V, Pekin D, Benhaim L, et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59(12):1722–1731. | ||
Klein AM, Mazutis L, Akartuna I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–1201. | ||
Ferraro D, Champ J, Teste B, et al. Microfluidic platform combining droplets and magnetic tweezers: application to HER2 expression in cancer diagnosis. Sci Rep. 2016;6:25540. | ||
Zhang H, Wang X, Ma Q, Zhou Z, Fang J. Rapid detection of low-abundance K-ras mutation in stools of colorectal cancer patients using chip-based temperature gradient capillary electrophoresis. Lab Invest. 2011;91(5):788–798. | ||
Duberow DP, Brait M, Hoque MO, et al. High-performance detection of somatic D-loop mutation in urothelial cell carcinoma patients by polymorphism ratio sequencing. J Mol Med. 2016:94(9):1015–1024. | ||
Blazej RG, Paegel BM, Mathies RA. Polymorphism ratio sequencing: a new approach for single nucleotide polymorphism discovery and genotyping. Genome Res. 2003;13(2):287. | ||
Paegel BM, Emrich CA, Wedemayer GJ, Scherer JR, Mathies RA. High throughput DNA sequencing with a microfabricated 96-lane capillary array electrophoresis bioprocessor. Proc Natl Acad Sci. 2002;99(2):574–579. | ||
Ferguson BS, Buchsbaum SF, Wu T-T, et al. Genetic analysis of H1N1 influenza virus from throat swab samples in a microfluidic system for point-of-care diagnostics. J Am Chem Soc. 2011;133(23):9129–9135. | ||
Song Y, Wang Y, Qin L. A multistage volumetric bar chart chip for visualized quantification of DNA. J Am Chem Soc. 2013;135(45):16785–16788. | ||
Heo HY, Chung S, Kim YT, Kim DH, Seo TS. A valveless rotary microfluidic device for multiplex point mutation identification based on ligation-rolling circle amplification. Biosens Bioelectron. 2016;78:140–146. | ||
Aebersold R, Anderson L, Caprioli R, Druker B, Hartwell L, Smith R. Perspective: a program to improve protein biomarker discovery for cancer. J Proteome Res. 2005;4(4):1104–1109. | ||
Laurell T, Marko-Varga G. Miniaturisation is mandatory unravelling the human proteome. Proteomics. 2002;2(4):345–351. | ||
Opitz CA, Kulke M, Leake MC, et al. Damped elastic recoil of the titin spring in myofibrils of human myocardium. Proc Natl Acad Sci U S A. 2003;100(22):12688. | ||
Peoples MC, Karnes HT. Microfluidic immunoaffinity separations for bioanalysis. J Chromatogr B. 2008;866(1–2):14–25. | ||
Chikkaveeraiah BV, Mani V, Patel V, Gutkind JS, Rusling JF. Microfluidic electrochemical immunoarray for ultrasensitive detection of two cancer biomarker proteins in serum. Biosens Bioelectron. 2011;26(11):4477–4483. | ||
Malhotra R, Patel V, Chikkaveeraiah BV, et al. Ultrasensitive detection of cancer biomarkers in the clinic by use of a nanostructured microfluidic array. Anal Chem. 2012;84(14):6249–6255. | ||
Crowther JR. The ELISA Guidebook. Berlin, Germany: Springer Science & Business Media; 2000. | ||
Chin CD, Laksanasopin T, Cheung YK, et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat Med. 2011;17(8):1015–1019. | ||
Park J, Sunkara V, Kim T-H, Hwang H, Cho Y-K. Lab-on-a-disc for fully integrated multiplex immunoassays. Anal Chem. 2012;84(5):2133–2140. | ||
Li Y, Xuan J, Song Y, et al. Nanoporous glass integrated in volumetric bar-chart chip for point-of-care diagnostics of non-small cell lung cancer. ACS Nano. 2016;10(1):1640–1647. | ||
Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew Chem Int Ed. 2007;46(8):1318–1320. | ||
Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Diagnostics for the developing world: microfluidic paper-based analytical devices. Anal Chem. 2010;82(1):3–10. | ||
Wang P, Ge L, Yan M, Song X, Ge S, Yu J. Paper-based three-dimensional electrochemical immunodevice based on multi-walled carbon nanotubes functionalized paper for sensitive point-of-care testing. Biosens Bioelectron. 2012;32(1):238–243. | ||
Wu Y, Xue P, Kang Y, Hui KM. Paper-based microfluidic electrochemical immunodevice integrated with nanobioprobes onto graphene film for ultrasensitive multiplexed detection of cancer biomarkers. Anal Chem. 2013;85(18):8661–8668. | ||
Xu S, Dong B, Zhou D, et al. Paper-based upconversion fluorescence resonance energy transfer biosensor for sensitive detection of multiple cancer biomarkers. Sci Rep. 2016;6:23406. | ||
Chen Y-H, Kuo Z-K, Cheng C-M. Paper – a potential platform in pharmaceutical development. Trends Biotechnol. 2015;33(1):4–9. | ||
Xu Y, Liu M, Kong N, Liu J. Lab-on-paper micro- and nano-analytical devices: fabrication, modification, detection and emerging applications. Microchim Acta. 2016;183(5):1521–1542. | ||
Xia Y, Si J, Li Z. Fabrication techniques for microfluidic paper-based analytical devices and their applications for biological testing: a review. Biosens Bioelectron. 2016;77:774–789. | ||
Huang B, Huang C, Liu P, Wang F, Na N, Ouyang J. Fast haptoglobin phenotyping based on microchip electrophoresis. Talanta. 2011;85(1):333–338. | ||
Kalish H, Phillips TM. Assessment of chemokine profiles in human skin biopsies by an immunoaffinity capillary electrophoresis chip. Methods. 2012;56(2):198–203. | ||
Lin X, Chen Q, Liu W, et al. Assay of multiplex proteins from cell metabolism based on tunable aptamer and microchip electrophoresis. Biosens Bioelectron. 2015;63:105–111. | ||
Nahavandi S, Baratchi S, Soffe R, et al. Microfluidic platforms for biomarker analysis. Lab Chip. 2014;14(9):1496–1514. | ||
Pagaduan JV, Sahore V, Woolley AT. Applications of microfluidics and microchip electrophoresis for potential clinical biomarker analysis. Anal Bioanal Chem. 2015;407(23):6911–6922. | ||
Sanjay ST, Fu G, Dou M, et al. Biomarker detection for disease diagnosis using cost-effective microfluidic platforms. Analyst. 2015;140(21):7062–7081. | ||
Kost GJ, Tran NK, Louie RF. Point-of-care testing: principles, practice, and critical-emergency-disaster medicine. In: Meyers RA, editor. Encyclopedia of Analytical Chemistry. John Wiley & Sons Ltd: Chichester, UK; 2008:1–45. | ||
Edited on behalf of the National Institute of Biomedical Imaging and Bioengineering/National Heart L, Price CP, Kricka LJ. Improving healthcare accessibility through point-of-care technologies. Clin Chem. 2007;53(9):1665–1675. | ||
Dhawan AP, Heetderks WJ, Pavel M, et al. Current and future challenges in point-of-care technologies: a paradigm-shift in affordable global healthcare with personalized and preventive medicine. IEEE J Transl Eng Health Med. 2015;3:1–10. | ||
ITU. ICT facts and figures – the world in 2015. Available from: http://www.itu.int/en/ITU-D/Statistics/Pages/facts/default.aspx. Accessed June 2, 2016. | ||
Wang S, Zhao X, Khimji I, et al. Integration of cell phone imaging with microchip ELISA to detect ovarian cancer HE4 biomarker in urine at the point-of-care. Lab Chip. 2011;11(20):3411–3418. | ||
Mancuso M, Cesarman E, Erickson D. Detection of Kaposi’s sarcoma associated herpesvirus nucleic acids using a smartphone accessory. Lab Chip. 2014;14(19):3809–3816. | ||
Priye A, Wong S, Bi Y, et al. Lab-on-a-drone: toward pinpoint deployment of smartphone-enabled nucleic acid-based diagnostics for mobile health care. Anal Chem. 2016;88(9):4651–4660. | ||
Contreras-Naranjo JC, Wei Q, Ozcan A. Mobile phone-based microscopy, sensing, and diagnostics. IEEE J Sel Top Quantum Electron. 2016;22(3):392–405. | ||
Hu J, Cui X, Gong Y, et al. Portable microfluidic and smartphone-based devices for monitoring of cardiovascular diseases at the point of care. Biotechnol Adv. 2016;34(3):305–320. | ||
Lee S, Srinivasan B, Vemulapati S, Mehta S, Erickson D. Personalized nutrition diagnostics at the point-of-need. Lab Chip. 2016;16(13):2408–2417. | ||
Molecular diagnostics market size | industry report, 2024. Available from: http://www.grandviewresearch.com/industry-analysis/molecular-diagnostics-market. Accessed November 28, 2016. | ||
Chin CD, Linder V, Sia SK. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip. 2012;12(12):2118–2134. | ||
Tay A, Pavesi A, Yazdi SR, Lim CT, Warkiani ME. Advances in microfluidics in combating infectious diseases. Biotechnol Adv. 2016;34(4):404–421. | ||
Becker H. Chips, money, industry, education and the “killer application.” Lab Chip. 2009;9(12):1659–1660. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
