Back to Journals » Infection and Drug Resistance » Volume 13
Microbiological Profile and Drug-Resistance Pattern of Pathogens Among Patients Who Visited the University of Gondar Comprehensive Specialized Hospital, Ethiopia
Authors Adane A, Belay G , Tamirat KS
Received 17 October 2020
Accepted for publication 3 December 2020
Published 14 December 2020 Volume 2020:13 Pages 4449—4458
DOI https://doi.org/10.2147/IDR.S287276
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Aynishet Adane,1 Gizeaddis Belay,2 Koku Sisay Tamirat3
1Department of Internal Medicine, School of Medicine, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia; 2Hospital Microbiology Department, University of Gondar Comprehensive Specialized Hospital, Gondar; 3Department of Epidemiology and Biostatistics, Institute of Public Health, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia
Correspondence: Koku Sisay Tamirat Email [email protected]
Background: The emergence of antimicrobial resistance (AMR) is a public health threat in developing countries including Ethiopia; and there is a paucity of information regarding antimicrobial resistance patterns of commonly isolated pathogens, particularly in the study area. Hence, this study aimed to assess the microbiological profiles and resistance patterns of pathogens among patients who visited a tertiary hospital in the study setting.
Methods: This study was based on secondary data sources from the hospital microbiology database and culture reports between September 2019 and August 2020 at the University of Gondar comprehensive specialized hospitals, Ethiopia. Data about socio-demographic characteristics and clinical parameters, types of specimens collected, culture results, and antibiotic resistance pattern were collected manually by using a data abstraction format from the department of clinical bacteriology registration book and electronic database.
Results: A total of 5328 culture results were included in the final analysis. Bacterial growth was documented only in 803 (15.1%) samples. From the positive culture results documented, the highest positivity rate was reported from abscess (47.8%) followed by blood (26.2%) and urine (15.1%) samples. Among the bacterial isolates S. aureus (32.5%), Klebsiella species (17.9%), E. coli (14.8%) and Streptococcus species (7.4%) were the commonly identified organisms. Of the 803 bacterial isolates, about 672 (83.6%) isolates were resistant to at least one antibiotic and 19.7% isolates were MDR.
Conclusion: This study showed that Staphylococcus aureus, Klebsiella pneumonia species, and Escherichia coli were the commonest isolated pathogens. Antimicrobial resistance among common isolates was high for most routinely used antibiotics, and some reserved drugs like carbapenems and fourth-generation cephalosporin. Thus, this study may have implications on patient management, drug procurement, local treatment guideline development, and rational use of antibiotics. Furthermore, this finding could also help to facilitate the implementation of antimicrobial stewardship and infection prevention and control interventions within the hospital.
Keywords: microbiological profile, antimicrobial resistance, Ethiopia
Background
Infectious diseases are the leading cause of morbidity and mortality and the major public health concern in developing countries.1 Different pathogens are responsible for infectious diseases that cause morbidity and mortality and highly resistant to commonly used antibiotics. Moreover, antibiotic resistance (AR) is a global health threat and the current agenda of the World health organization (WHO) and other international organizations. In 2015 WHO released the Global Action Plan (GAP) on antimicrobial resistance which outlines five different objectives that aimed at ensuring treatment and prevention of infectious diseases with quality-assured, safe, and effective medicines.2
Antimicrobial resistance (AMR) threatens the effective prevention and treatment of an ever-increasing range of infections caused by bacteria, parasites, viruses, and fungi strains.3–6 Irrational use of antibiotics, suboptimal quality drugs, and empirical treatment of infections without adequate laboratory evidence contributed to the occurrence of antimicrobial resistance.7–10 Evidences showed that 25,000 annual deaths, and 2.5 million extra days of hospital stays are attributed to antimicrobial resistance. Irrational use of antibiotics, poor drug quality, and the smuggling of substandard drugs are the major drivers of rising antimicrobial resistance in developing countries.11–13 Besides, poor infection prevention control practices in the health facilities also contributed to the increased occurrence of antimicrobial resistance. In addition, the transmission of healthcare-associated infections in hospitals is also responsible to the widespread of antimicrobial-resistant germs.
Recently, a group of bacteria has been described by the acronym of ESKAPE which refers Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species predominantly causes most of the nosocomial infections in healthcare settings.7,8,11–14
Developing countries are characterized by poor health facility infrastructures and overwhelmed by high burdens of infectious diseases like tuberculosis and HIV/AIDS. Additionally, there is a lack of information about the bacteriologic profile and antimicrobial susceptibility patterns in health facilities of resource-limited settings including the study area. Therefore, this study aimed to assess the microbiological profile and resistance pattern of pathogens isolated from patients who visited the University of Gondar comprehensive specialized hospital.
Method
Study Design, Setting, and Period
This study was a descriptive cross-sectional based on secondary data from the hospital microbiology database and culture reports between September 2019 and August 2020 at the University of Gondar Comprehensive Specialized Hospitals, Ethiopia. The hospital is one of the tertiary hospitals in Northwest Ethiopia and serves more than seven million people in the catchment area. The University hospital has separated microbiological and tuberculosis culture units; and a functioning antimicrobial stewardship and infection prevention and control committee. As an average, close to 6000 cultures are being performed in the hospital annually.
Data Collection and Specimen Processing
Data about socio-demographic characteristics and clinical parameters such as age, sex, names of wards and departments, types of samples collected, culture results, and antibiotic susceptibility pattern were collected manually by using a pre-prepared data abstraction format from secondary data sources obtained from the department of clinical bacteriology registration book and electronic database whereby all the necessary clinical and microbiology-related information were recorded.
After a request by the treating physician, different culture samples were collected from respective units of the hospital. The standard operating procedures of collection, storage, and transport of samples were implemented. After culture samples get collected by the microbiology unit; culture Media were tested for sterility and performance according to CLSI standard procedures.15
A drop of different specimens was placed on chocolate, blood, and MacConkey agar plates; and stroked over the culture media. The chocolate and blood agar plates were incubated in a CO2 enriched atmosphere at 35–37°C for up to 72 hours, on the other hand, MacConkey agar plates were incubated aerobically. After overnight incubation, culture media were checked for growth manually; and whenever there was any growth after overnight incubation, the definitive biochemical identification of the organism and antimicrobial susceptibility test was performed. Colony morphology, hemolytic pattern, Gram stain, and biochemical tests including lactose fermentation, lysine, citrate and urea utilization, motility, and gas production were used to identify types of bacteria.16
Susceptibilities to common antibiotics were determined by the Kirby-Bauer disc diffusion method.17 The drugs tested for both gram-negative and gram-positive bacteria were: Amoxicillin/clavulanic acid (10µg), Ciprofloxacin (10µg), Gentamicin (10µg), Trimethoprim/sulfamethoxazole (10µg), chloramphenicol (10µg), cefotaxime (30µg), Cefepime (10µg) and ceftriaxone (30µg). Moreover, penicillin (10µg), Tetracycline (10µg), cefoxitin (10µg), clindamycin (10µg), vancomycin (30 µg), and erythromycin (15µg) drugs were exclusively used to test gram-positive bacteria whereas, Ampicillin (10µg), ceftazidime (30µg), Norfloxacin (30 µg), Amikacin (10µg), Imipenem (10µg) and meropenem (10µg) were used for gram-negative bacterial isolates. The zones of inhibition were measured and compared with the National Committee for Clinical Laboratory Standards (NCCLS) guidelines.18 International reference bacteria strains: Escherichia coli (ATCC 25922) and Staphylococcus aureus (ATCC 25923) were used as controls.19
For this study, intermediate antimicrobial susceptibility was considered as resistance and multi-drug resistance (MDR) was defined as the resistance of bacterial isolates to at least two or more different antibiotics tested.
Data Analysis and Interpretation
The data were checked for inconsistencies, coding errors, completeness, clarity, and missing values before entry. The data entry was done using Epi-data version 4.64.0 and then it was exported to SPSS version 22 for further analysis. Descriptive statistics were carried out and presented using tables, text, and graphs. In addition, cross-tabulation of culture results with socio-demographic variables was done to check if there was any association with culture positivity. Thus, variables having less than 0.05 P-value were considered to have a statistically significant association.
Result
Socio-Demographic Characteristics
A total of 5340 culture samples were collected during the study period, of which 12 samples were excluded due to incomplete information. As a result, the final samples analyzed for this study were 5328, close to 60% of the patient samples included were from male patients. More than half (55%) patients aged above 15 years; and about 8.3% of them were neonates below the age of 28 days. Additionally, the majority of culture samples were obtained from the pediatric emergency unit, adult emergency unit, and medical wards contributing about 30.3%, 24%, and 18.2%, respectively (Table 1).
 |
Table 1 Socio-Demographic Characteristics of Patients Whose Samples Were Included (N=5328) |
Microbiological Profile of Culture Isolates
Out of the total 5328 cultures performed, more than a quarter (27.4%) of the samples constituted blood cultures followed by CSF (25.5%) and other body fluids (16.3%) (Figure 1). From all the cultures analyzed during one year period, bacterial growth was documented only in 803 (15.1%) samples and contaminants were also isolated among 132 (2.5%) samples. Culture samples collected from the pediatric ward accounted for the highest positivity result (27.9%) followed by samples collected from medical wards (19.3%), surgical wards (17.8%), and ICU (10.2%) respectively. More than half (57.3%) of positive culture results were obtained from samples collected from the age group of above 15 years. From the positive culture results documented, the highest positivity rate was obtained from abscess (47.8%) followed by blood (26.2%) and urine (15.1%) samples (Figure 2). Findings also revealed that culture positivity had statistically significant association with the types of samples collected (P=0.0001) and place of sample collection (P=0.0001). However, no statistically significant association was noted with the other variables including, age (P= 0.705) and sex of patients (P= 0.475). Among the bacterial isolates: S. aureus (32.5%), Klebsiella species (17.9%), E.coli (14.8%), and Streptococcus species (7.4%) were the most commonly identified organisms (Figure 3) and (Tables 2 and 3).
 |
Table 2 Bacterial Isolates from Clinical Samples (N=5328) |
 |
Table 3 Bacteriological Isolates by Socio-Demographics and Hospital Departments (Place of Specimen Collection) |
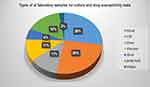 |
Figure 1 A pie chart showing the types of laboratory samples for culture and drug susceptibility tests. |
 |
Figure 2 Culture positivity of common samples and isolates at the University of Gondar comprehensive specialized hospital. |
 |
Figure 3 Graph showing the bacterial isolates by hospital departments. |
Antimicrobial Resistance Pattern
Of the 803 culture isolates about six hundred seventy-two (83.6%) isolates were resistant to at least one antibiotic and 19.7% isolates were multidrug-resistant (MDR). More specifically, 34.8%, 28.7%, 26.8%, and 22.6% of isolated S.aureus species were resistant to Penicillin, Erythromycin, TTC, and clindamycin, respectively.
From the gram-negative isolates, Klebsiella species showed resistance to ceftazidime (59%), Cefuroxime (44.4%), Ceftriaxone (38.2%), ciprofloxacin (45.1%), and trimethoprim or sulfamethoxazole (SXT) (58.3%). On the other hand, out of the 119 E.coli isolates, most were resistant to widely used antimicrobials like fluoroquinolones and cephalosporin including ciprofloxacin (47.9%), ceftazidime (50.4%), ceftriaxone (39.5%), and cefuroxime (35.3%).
In addition, about 47.3%, 43.7%, 25%, 25% of Pseudomonas species were resistant to trimethoprim or sulfamethoxazole (SXT), ceftazidime, ceftriaxone, and amoxicillin/clavulanic acid antibiotics, respectively. Notably, out of all gram-negative isolates, only three hundred twenty-two gram-negative pathogens had been tested for Carbapenems susceptibility from which 39 (12.1%) of the bacteria isolates were resistant to either imipenem or meropenem. The details of antimicrobial resistance patterns of isolates are presented in Tables 4–6.
 |
Table 4 Drug Susceptibility Test Results of Bacteriological Isolates at the University of Gondar Comprehensive Specialized Hospital |
 |
Table 5 Drug Susceptibility Test Results of Bacteriological Isolates at the University of Gondar Comprehensive Specialized Hospital (Continued) |
 |
Table 6 Drug Susceptibility Test Results of Bacteriological Isolates at the University of Gondar Comprehensive Specialized Hospital for Selected Antibiotics |
Discussion
This study showed that more than half (55.8%) of patients aged above 15 years and 8.3% of the patients were neonates. Blood (28%), Cerebrospinal fluid (CSF) (26%), and body fluid (16%) were the most frequently collected samples. From 5328 patient cultures during one year period, bacterial growth was documented only in 803 (15.1%) culture samples. The rate of culture positivity in this study was comparable with other previous findings reported from Jimma medical center (15.8%).20 However, this study finding was lower than other similar studies conducted in Ethiopia, particularly in Addis Ababa (27.9%)21 and Dessie (22.7%);22 and a study done in India (22.5%).23 In addition, this study showed that about 132 (2.5%) samples grew contaminants. The rate of contamination in this study appeared to be within acceptable ranges recommended by WHO.24 Regarding the place of collection, culture samples collected from the pediatric ward accounted for the highest positivity result (27.9%) followed by medical wards (19.3%), surgical wards (17.8%) and ICU (10.2%), adult emergency (9.1%) and neonatal ICU (7.5%). As depicted in Figure 2 Klebsiella species was the most common isolate from CSF (41.4%) and Urine (27.3%) samples, whereas, S. aureus was the commonest isolate from Abscess (47%) and Blood (26.3%) samples. These findings were consistent with the studies mentioned above.11,25 Regarding the rate of culture positivity related to the type of samples, the highest percentage of bacterial growth was documented from abscess samples (47.8%) followed by blood (26.2%) and urine (15.1%). Overall, S. aureus (32.5%), Klebsiella species (17.9%), E.coli (14.8%), and Streptococcus species (7.4%) were the commonest identified organisms. This finding was consistent with systematic review findings in Africa and similar studies conducted in Ethiopia.5,7,11 Regarding the specific age groups, Klebsiella species (46.7%) was the commonest bacteria isolated among the neonatal age group followed by S. aureus (26.7%), whereas, S. aureus (26.9%), E.coli (18.3%), and Klebsiella species (16.1%) were the most common isolates in adults. This finding was consistent with the previously mentioned studies.5,25,26 This finding might suggest that the source of infections to neonates might be the mother or acquired in the hospitals during or after delivery as a result of poor infection prevention and control activities within the hospital.
Regarding the resistance pattern, out of all culture isolates, about six hundred seventy-two (83.6%) pathogens were resistant to at least one antibiotic. The S.aureus species isolates showed resistance to routinely prescribed antibiotics like: penicillin (34.8%), Erythromycin (28.7%), Tetracycline (26.8%), clindamycin (22.6%), SXT (23.4%) and Cefoxitin (17.2%). This current study also revealed that gram-negative pathogens like pseudomonas, Klebsiella, and E.coli were highly resistant to commonly used antibiotics such as cephalosporin, fluoroquinolones, aminoglycosides, and trimethoprim or sulfamethoxazole (SXT). This finding was consistent with the finding from the antimicrobial resistance surveillance study from Ethiopia.25
In addition, this study also showed that Acinetobacter species identified were commonly resistant to antibiotics like Trimethoprim/Sulfamethoxazole (SXT) (55.7%), Ceftazidime (53.8%), Gentamycin (50%), and Cefuroxime (46.1%).
Generally, our study finding revealed that most isolated pathogens were highly resistant to the commonly utilized antibiotics in the hospital which may directly affect the quality of patient care, efficiency of the hospital, and cost of patient treatment. Hence, the development of evidence-based local guidelines, modification of treatment protocols, and leveraging antimicrobial stewardship and IPC interventions are relevant to mitigate the high burden of antimicrobial resistance in the study setting.25,26
Limitation of the Study
The retrospective nature of the study has led missing of some clinical and laboratory variables; and most importantly, our study did not support the integration of microbiology findings with stewardship and IPC interventions; and was not well incorporated in the routine patient care. Shortage of some required materials and supplies in the hospital might harm the quality of culture result. For instance, susceptibility tests to some important antibiotics like Carbapenems, methicillin, and vancomycin were not consistently being performed in the hospital during the study period. Generally, only those antibiotics tested in the hospital laboratory were included in this study. Thus, it may not include all antibiotics used in routine clinical practice. Since the majority of the samples were collected from Internal medicine and Pediatrics departments, the finding of this study might not be representative of the whole units of the hospital.
Conclusion
This study showed that Staphylococcus aureus, Klebsiella pneumonia species, and Escherichia coli were the commonest isolated pathogens. Antimicrobial resistance among common isolates was high for routinely used antibiotics; particularly to cephalosporin, fluoroquinolones, aminoglycosides, and trimethoprim or sulfamethoxazole; and to some reserved drugs like Carbapenems and fourth-generation Cephalosporin.
In general, this study has just revealed an overview of common pathogens isolated in our hospital and their resistance profile which may guide clinicians, hospital stewardship and IPC committees, policymakers, and hospital managers for future measures. In addition, the findings of this study could also be used as a baseline by researchers for further prospective studies and interventions.
Abbreviations
CSF, cerebrospinal fluid; ICU, intensive care unit; IPC, infection prevention and control; MDR, multi-drug resistant; NCCLS, National Committee for Clinical Laboratory Standards; NICU, Neonatal Intensive Care Unit; TB, tuberculosis; OPD, outpatient department; trimethoprim/sulfamethoxazole (SXT) WHO, World Health Organization.
Data Sharing Statement
The datasets used during the current study are available from the corresponding author.
Ethical Considerations
Ethical clearance was obtained from the University of Gondar ethical review board. In addition, permission was also obtained from the hospital chief clinical director and microbiology unit head of the hospital. Patient identifiers were not used to ensure the privacy of study participants. All information obtained during this study was kept confidential and used merely for the research purpose. Regarding ethical issues, this study protocol was conducted in accordance with the World Medical Association (WMA) Declaration of Helsinki.
Consent for Publication
Not applicable as there is no image or other confidentiality related issues.
Acknowledgment
We would like to acknowledge data collectors, hospital administrators, and Microbiology department staff for their support.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
We did not receive external funds for this research.
Disclosure
The authors declared that they have no conflicts of interest for this work.
References
1. Asokan GV, Ramadhan T, Ahmed E, et al. WHO global priority pathogens list: a bibliometric analysis of Medline-PubMed for knowledge mobilization to infection prevention and control practices in Bahrain. Oman Med J. 2019;34(3):184. doi:10.5001/omj.2019.37
2. Mendelson M, Matsoso MP. The World Health Organization global action plan for antimicrobial resistance. S Afr Med J. 2015;105(5):325. doi:10.7196/SAMJ.9644
3. Bernabe KJ, Langendorf C, Ford N, et al. Antimicrobial resistance in West Africa: a systematic review and meta-analysis. Int J Antimicrob Agents. 2017;50(5):629–639. doi:10.1016/j.ijantimicag.2017.07.002
4. Beyene D, Bitew A, Fantew S, et al. Multidrug-resistant profile and prevalence of extended spectrum β-lactamase and carbapenemase production in fermentative Gram-negative bacilli recovered from patients and specimens referred to National Reference Laboratory, Addis Ababa, Ethiopia. PLoS One. 2019;14(9):e0222911. doi:10.1371/journal.pone.0222911
5. Dagnew M, Yismaw G, Gizachew M, et al. Bacterial profile and antimicrobial susceptibility pattern in septicemia suspected patients attending Gondar University Hospital, Northwest Ethiopia. BMC Res Notes. 2013;6(1):283. doi:10.1186/1756-0500-6-283
6. Moremi N, Claus H, Mshana SE. Antimicrobial resistance pattern: a report of microbiological cultures at a tertiary hospital in Tanzania. BMC Infect Dis. 2016;16(1):756. doi:10.1186/s12879-016-2082-1
7. Ntirenganya C, Muvunyi CM, Manzi O, et al. High prevalence of antimicrobial resistance among common bacterial isolates in a tertiary healthcare facility in Rwanda. Am J Trop Med Hyg. 2015;92(4):865–870. doi:10.4269/ajtmh.14-0607
8. Okoche D, Asiimwe BB, Katabazi FA, et al. Prevalence and characterization of carbapenem-resistant Enterobacteriaceae isolated from Mulago National Referral Hospital, Uganda. PLoS One. 2015;10(8):e0135745. doi:10.1371/journal.pone.0135745
9. Tadesse BT, Ashley EA, Ongarello S, et al. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis. 2017;17(1):616. doi:10.1186/s12879-017-2713-1
10. Wasihun AG, Wlekidan LN, Gebremariam SA, et al. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle Hospital, Northern Ethiopia. Springerplus. 2015;4(1):314. doi:10.1186/s40064-015-1056-x
11. Carroll M, Rangaiahagari A, Musabeyezu E, et al. Five-year antimicrobial susceptibility trends among bacterial isolates from King Faisal Hospital Tertiary Health-Care Facility in Kigali, Rwanda. Clin Ther. 2017;39(8):e4–e5. doi:10.1016/j.clinthera.2017.05.017
12. Le Doare K, Bielicki J, Heath PT, et al. Systematic review of antibiotic resistance rates among gram-negative bacteria in children with sepsis in resource-limited countries. J Pediatric Infect Dis Soc. 2015;4(1):11–20. doi:10.1093/jpids/piu014
13. Thaden JT, Li Y, Ruffin F, et al. Increased costs associated with bloodstream infections caused by multidrug-resistant gram-negative bacteria are due primarily to patients with hospital-acquired infections. Antimicrob Agents Chemother. 2017;61(3). doi:10.1128/AAC.01709-16.
14. Maina D, Omuse G, Revathi G, et al. Spectrum of microbial diseases and resistance patterns at a private teaching hospital in Kenya: implications for clinical practice. PLoS One. 2016;11(1):e0147659. doi:10.1371/journal.pone.0147659
15. Weinstein MP, Lewis JS, Kraft CS. The clinical and laboratory standards institute subcommittee on antimicrobial susceptibility testing: background, organization, functions, and processes. J Clin Microbiol. 2020;58(3). doi:10.1128/JCM.01864-19
16. Cheesbrough M. District Laboratory Practice in Tropical Countries, Part 2. Cambridge university press; 2006.
17. Wayne P Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing; 2011.
18. Wayne P. National committee for clinical laboratory standards. In: Performance Standards for Antimicrobial Disc Susceptibility Testing. Vol. 12. 2002:01–53.
19. Clinical and L.S. Institute. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
20. Kumalo A, et al. Bacterial profile of adult sepsis and their antimicrobial susceptibility pattern at Jimma University specialized hospital, south West Ethiopia. Health Sci J. 2016;10(2):1.
21. Negussie A, et al. Bacteriological profile and antimicrobial susceptibility pattern of blood culture isolates among septicemia suspected children in selected hospitals Addis Ababa, Ethiopia. Int J Biol Med Res. 2015;6(1):4709.
22. Kibret M, Abera B. Prevalence and antibiogram of bacterial isolates from urinary tract infections at Dessie Health Research Laboratory, Ethiopia. Asian Pac J Trop Biomed. 2014;4(2):164–168. doi:10.1016/S2221-1691(14)60226-4
23. Pal N, Sujatha R. Antimicrobial resistant pattern of blood culture isolates, among septicaemia suspected patients. Natl J Lab Med. 2016;5(1):17–21.
24. Roth A, Wiklund AE, Pålsson AS, et al. Reducing blood culture contamination by a simple informational intervention. J Clin Microbiol. 2010;48(12):4552–4558. doi:10.1128/JCM.00877-10
25. Ibrahim RA, Teshal AM, Dinku SF, et al. Antimicrobial resistance surveillance in Ethiopia: implementation experiences and lessons learned. Afr J Lab Med. 2018;7(2):1–4. doi:10.4102/ajlm.v7i2.770
26. Essack S, et al. Antimicrobial resistance in the WHO African region: current status and roadmap for action. J Public Health. 2017;39(1):8–13.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
