Back to Journals » Cancer Management and Research » Volume 12
METTL14 Overexpression Promotes Osteosarcoma Cell Apoptosis and Slows Tumor Progression via Caspase 3 Activation
Authors Liu Z, Liu N, Huang Z, Wang W
Received 30 September 2020
Accepted for publication 30 November 2020
Published 11 December 2020 Volume 2020:12 Pages 12759—12767
DOI https://doi.org/10.2147/CMAR.S284273
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
Ziwen Liu, Ning Liu, Zhipeng Huang, Wenbo Wang
Department of Orthopedics, The First Affiliated Hospital of Harbin Medical University, Harbin 150086, People’s Republic of China
Correspondence: Wenbo Wang
Department of Orthopedics, The First Affiliated Hospital of Harbin Medical University, Harbin 150001, People’s Republic of China
Email [email protected]
Ning Liu
Department of Orthopedics, The First Affiliated Hospital of Harbin Medical University, Harbin 150001, People’s Republic of China
Email [email protected]
Background: As a key enzyme of m6A methylation modification, methyltransferase-like 14 (METTL14) is involved in many physiological and pathophysiological processes. This study aims to explore the effect of METTL14 on the viability of osteosarcoma cells and explain the underlying molecular mechanism.
Methods: We detected the content of METTL14 in osteosarcoma tissue by qRT-PCR and Western blot. Experiments such as transwell, EdU, and CCK-8 have demonstrated the effect of METTL14 on osteosarcoma cell activity. In addition, the regulation of caspase-3 by METL14 was determined by Western blot. We used caspase-3 inhibitor to further reverse the effect of METTL14 on osteosarcoma cell apoptosis.
Results: We found that the expression of METTL14 in osteosarcoma cells was reduced compared with normal tissues. METTL14 overexpression significantly reduced the proliferation, migration, invasion and apoptosis of osteosarcoma cells. Inhibition of METL14 showed the opposite result. We have demonstrated that METTL14 finally achieves apoptosis by activating caspase-3.
Conclusion: We have demonstrated that METTL14 has effects on osteosarcoma cell proliferation, migration, and invasion and promotes cell apoptosis by activating caspase-3, which may become a potential therapeutic target for osteosarcoma.
Keywords: osteosarcoma, m6A methylation, METTL14
Introduction
Osteosarcoma (OS) is a malignant tumor with a high degree of malignancy and a poor prognosis, which tends to occur in adolescents.1 The current clinical treatment is mainly surgical treatment, supplemented by radiotherapy, chemotherapy and biological adjuvant therapy.2,3 However, due to many complications of surgical treatment, tumor cells are prone to multi-drug resistance and insensitivity to radiotherapy, and the 5-year survival rate of patients has not improved in recent years.4,5 Therefore, it is of great significance to explore new targeted therapies to improve the clinical outcome of OS patients.
6-methyladenosone (N6-methyladenosone, m6A), the methylation that occurs on the sixth N of adenylate (A) in RNA, is the most abundant epitranscriptomic modification in eukaryotic mRNA.6 m6A RNA methylation is a dynamic and reversible modification process. From catalytic formation to functional realization, it is mainly affected by regulation of the m6A methyltransferase complex (METTL3, METTL14 and WTAP), m6A demethylase (FTO, ALKBH5) and m6A reader proteins.7–9 Studies have found that m6A may affect a series of life processes such as development, immune regulation, tumor progression and stem cell differentiation.6,10 Several related studies have reported on m6A-related modified enzymes and their role in the occurrence and development of different human tumors.11 m6A-related modifying enzymes regulate the expression of proto-oncogenes and tumor suppressor genes by affecting specific RNA molecules or the overall RNA m6A modification levels.8,12 In addition, other mechanisms for changing m6A modifications include the gain or loss of specific m6A modifications caused by point mutations.13
As a key component of the m6A methyltransferase complex, METTL14 plays an important role in the development of cancer cells.14 Weng et al15 found that METTL14 modulates mRNA targets of acute myeloid leukemia (AML) (for example, MYB and MYC) to exert carcinogenic effects. Ma et al16 reported that METTL14 inhibits the metastasis of liver cancer cells by regulating the m6A methylation modification of microRNA126. Chen et al17 and other studies have shown that overexpression of METTL14 can inhibit the growth and metastasis of colorectal cancer cells. However, the effect of METTL14 on osteosarcoma cells has not been examined. This study aims to explore the role of METTL14 in the occurrence and development of osteosarcoma cells and provide new ideas for the clinical treatment of osteosarcoma.
In this study, we observed changes in osteosarcoma cells by regulating the expression of METTL14. To that end, we found that METTL14 can inhibit the proliferation, migration and invasion of tumor cells, and promote tumor cell apoptosis.
Materials and Methods
Cell Culture and Treatment
Human osteosarcoma cell lines U2OS and 143B were purchased from Shanghai Institutes of Biological Sciences, Chinese Academy of Sciences (Shanghai, China). U2OS was cultured in Dulbecco`s modified Eagle medium (DMEM) (Life Technologies Corporation, California, USA) supplemented with 10% fetal bovine serum (FBS) (Biological Industries, Israel) at 37°C under 5% CO2. 143B cells were cultured in RPMI 1640 basic medium (Cat# 11–875-093; Gibco, USA) at 37°C under 5% CO2.
qRT-PCR
Total RNA was extracted using TRIzol reagent (Life technologies Corporation) and then 500 ng total RNA was reverse transcribed to cDNA using a High Capacity cDNA Reverse Transcription Kit (Cat# 00676299; Thermo Fisher Scientific, Waltham, USA). Quantitative reverse transcription PCR analysis was performed with 1 μL cDNA using SYBR Green PCR Master (Roche) in a 7500 Fast Real-Time instrument (Applied Biosystems, Foster City, CA, United States). GAPDH was used as an endogenous control. Reactions were run in triplicate. The primer pairs used in our PCR analysis are as follows (Forward/Reverse primer sequence (5`-3`)).
METTL14-F: GTCTTAGTCTTCCCAGGATTGTTT
METTL14-R: AATTGATGAGATTGCAGCACC
GAPDH-F: AGCCACATCGCTCAGACAC
GAPDH-R: GCCCAATACGACCAAATCC
Western Blot
Cells were lysed in cell lysis buffer (Cat# P0013B; Beyotime Biotechnology, Shanghai, China) supplemented with PMSF protease inhibitor on ice for 1 h, followed by centrifuging at 13,500 g at 4°C for 15 min. Protein concentration was measured using a BCA Protein Assay Kit (Cat# P0010S; Beyotime Biotechnology) following the manufacturer’s instructions. Protein samples (50 µg) were separated on a polyacrylamide gel and transferred to a nitrocellulose membrane and then blocked with 10% fat-free dry milk at room temperature for 1 h. Then, the membrane was incubated with a rabbit anti-METTL14 antibody (1:1000; Cat# 51104S; Cell Signaling Technology), a rabbit anti-caspase-3 antibody (1:1000; Cat# 9662S; Cell Signaling Technology), and a mouse anti-GAPDH antibody (1:500; Cat# abs830030; Absin) at 4°C overnight. Then, the membrane was incubated with monoclonal anti-rabbit IgG (1:5000; Cat# ab97051; Abcam) or monoclonal anti-mouse IgG (1:5000; Cat# ab6789; Abcam) at room temperature for 1 h. Western blot bands were imaged with Odyssey CLx and quantified with LI-COR Image Studio Software (LI-COR Biosciences, Lincoln, NE, USA).
Plasmid Transfection
The METTL14-carrying plasmid for overexpression was constructed by Cyagen (Suzhou, China). Cells were plated in 6-well culture plates and transfected with 1 μg plasmid using LipofectamineTM 3000 Transfection Reagent (Cat# L3000008; Cell Signaling Technology) according to the manufacturer’s protocols. Cells were collected 24 h after transfection.
Cell Counting Kit (CCK)-8 Assay
Cell Counting Kit (CCK)-8 assay was used to measure cell proliferation ability. 1000 cells/well were seeded in 96-well plates. At 0, 24, 48 and 72 h, 100 μL fresh medium containing 10 μL CCK-8 (Cat# HY-K0301, MCE) was added into each well to replace the original medium. A spectrophotometer was used to measure the absorbance of each well (Thermo Scientific, Waltham, MA, USA).
Ethynyl-2-Deoxyuridine (EdU) Staining Assay
An EdU Apollo DNA in vitro kit (Ribobio, Guangzhou, China) was used to detect cell proliferation ability. Cells were plated into 12-well culture plates at a density of 4.0×104. Briefly, cells were incubated with 30 μM EdU at 37°C for 90 min and then fixed with 4% paraformaldehyde (m/v) for 30 min. After permeabilized in 0.5% Triton X-100, the Apollo staining solution was added into the cell culture medium for 30 min away from light. Finally, the cells were incubated with 20 μg/mL 4ʹ,6-diamidino-2-phenylindole (DAPI) for 10 min. The EdU index (%) is the average ratio of the number of EdU-positive cells over total cells in five randomly selected areas under the confocal laser scanning microscope (FV10i).
Invasion Assay
Twenty-four-millimeter transwell chambers (Corning #3412, USA) were used to detect cell invasive abilities, and the assay was performed according to the manufacturer’s protocols. A total of 1 × 104 cells infected with plasmid were resuspended in 200 μL serum-free DMEM medium and seeded in the upper chamber. DMEM medium containing 10% FBS was added into the lower chamber. After 24 h, cells migrated through the membrane and were stained with 0.1% crystal violet (Beyotime Biotechnology, China) for 15 min and counted using light microscopy (ECLIPSE TS100, Nikon).
Migration Assay
Cells were plated into 6-well culture plates at a density of 2.5 × 105 cells/mL. When the confluence of cells reached 70%, a wound was created by scraping the cells with a 200-μL pipette tip. Cells were washed with PBS and then transfected with plasmid. Images were captured at 0, 24 and 48 h after wounding by standard light microscopy (ECLIPSE TS100, Nikon, Japan). The wound area was measured using ImageJ software (National Institutes of Health (NIH), USA).
Colony-Formation Assay
Cells transfected with the targeted plasmid were seeded in a six-well plate at a concentration of 1500 cells per well and cultured at 37°C under 5% CO2. Two weeks later, cells were fixed and stained separately with 100% methanol and 0.1% crystal violet for 20 min. Colonies were air-dried and counted. The experiments were repeated three times.
Propidium Iodide (PI)/Hoechst 33,342 Staining
PI/Hoechst 33,342 staining was performed using a Hoechst 33,342/PI Double Staining Kit (Solarbio Science, Beijing China). The cells were evenly grown on the bottom of the culture dish, fixed with 4% paraformaldehyde at 37°C for 15 min, and stained with the dye solution at room temperature and away from light for 30 min. Finally, images of the staining were captured by a confocal laser scanning microscope (FV10i, Olympus, Tokyo, Japan).
Statistical Analysis
Data are expressed as mean ± SEM. Statistical analysis was performed using GraphPad Prism7 software and analyzed by Student’s t-test (two-tailed). All experiments were independently repeated at least three times. *P < 0.05; **P < 0.01; ***P < 0.001.
Results
METTL14 Expression is Reduced in Osteosarcoma Tissues
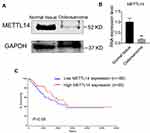
To clarify the relationship between METTL14 and osteosarcoma, we determined the content of METTL14 in normal tissues and osteosarcoma tissues by Western blot. The results showed that the content of METTL14 protein in normal tissues is much higher than that of the osteosarcoma patient group (Figure 1A). In addition, the expression of METTL14 RNA in osteosarcoma tissue was lower than that of normal tissues (Figure 1B). Furthermore, the survival analysis from the Cancer Genome Atlas (TCGA) data set (http://www.oncolnc.org/) showed that patients with high METTL14 expression have a higher early survival rate. On the contrary, patients with low METTL14 expression have poor early survival rates (Figure 1C).
METTL14 Overexpression Can Inhibit Cell Viability and Proliferation of Osteosarcoma Cells
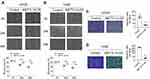
To evaluate the effect of METTL14 on the proliferation of osteosarcoma cells, we chose to overexpress METTL14 in the human osteosarcoma cell lines U2OS and 143B. EDU staining results showed that overexpression of METTL14 could significantly inhibit the proliferation of osteosarcoma cells (Figure 2E and F). Additionally, through the Cell Counting Kit-8 (CCK8) experiment to detect cell proliferation capacity, in the two cell lines, the OD value of the overexpression group was significantly lower than that of the control group at 24 hours, 48 hours and 72 hours (Figure 2A and B). Moreover, METTL14 could significantly inhibit the activity of osteosarcoma cells. In addition, cell clone formation experiments showed that when METTL14 was overexpressed, the number of osteosarcoma cell clones decreased (Figure 2C and D).
METTL14 Overexpression Can Inhibit the Migration and Invasion of Osteosarcoma Cells
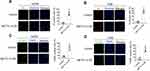
The cell scratch experiment was used to explore the effect of METTL14 on the migration ability of osteosarcoma cells. The results showed that the migration speed of the overexpression group at 24 hours and 48 hours in the two cell lines was significantly lower than that of the control group (Figure 3A and B). This demonstrates that METTL14 can inhibit the migration ability of osteosarcoma cells. A transwell invasion experiment was used to further study the influence of METTL14 on the invasion ability of osteosarcoma cells. Compared with the control group, the invasive cells in the overexpression group in the two cell lines were significantly reduced, indicating that METTL14 can inhibit the invasive ability of osteosarcoma cells (Figure 3C and D).
METTL14 Overexpression Can Promote Osteosarcoma Cell Apoptosis
To clarify the effect of METTL14 on osteosarcoma cell apoptosis, we performed PI staining in two cell lines. The results showed that the amount of apoptosis in the overexpression group in the two cell lines was significantly greater than that of the control group, and the overexpression of METTL14 could promote the apoptosis of osteosarcoma cells (Figure 4A and B). To further verify this conclusion, we conducted a TUNEL staining experiment. These results also confirmed that overexpression of METTL14 can promote cell apoptosis (Figure 4C and D).
Anti-Tumor Property of METTL14 is Through Caspase 3 Cleavage
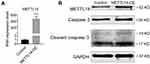
As we observed METTL14 could strongly induce cell apoptosis of osteosarcoma cell, we assessed the protein level of cleaved caspase 3, the most important terminal shear enzyme during cell apoptosis. As shown in Figure 5, 24 hours after transfection of METTL14 plasmids, the protein level of cleaved caspase 3 dramatically decreased along with the increase of METTL14 in U2OS cells (Figure 5A and B). To further confirm the effects of METTL14 in osteosarcoma cell viability was solely responsible for apoptosis, we combined METTL4 overexpression treatment with the general caspase inhibitor Z-VAD-FMK. As expected, combined treatment with Z-VAD-FMK significantly reverted the cleaved caspase-3 protein levels enhanced by METTL14 overexpression in U2OS cells (Figure 6A). Consistently, PI and TUNEL staining results showed that Z-VDA-FMK markedly attenuated destruction of METTL14 for cell activity in both U2OS and 143B cells (Figure 6B–E).
Discussion
As an important part of the encoder in the process of RNA methylation, METTL14 plays an important role in the occurrence and development of a variety of tumor cells. Osteosarcoma, a malignant tumor, has not been clinically effective in recent years to improve the 5-year survival rate of patients. At present, there is still a lack of research on the effect of METTL14 on osteosarcoma cells. Our experiments demonstrated that when METTL14 expression is enhanced, it can inhibit the activity of osteosarcoma cells and promote cell apoptosis. By examining the content of METTL14 in tissues, we found that METTL14 is positively correlated with the prognosis of patients with osteosarcoma. Therefore, from these experiments, we propose that downregulation of METTL14 may be a potential mechanism underlying osteosarcoma and can be used as a tumor suppressor gene, and overexpression of METTL14 can be used as a new alternative therapy for osteosarcoma.
Recent studies have shown that m6A methylation modifying enzymes are abnormally expressed in some malignant tumors. Li et al,18 found that in OS patients, especially in metastatic tissues, disorders involving m6A-related regulatory factors, such as the low expression of METTL3 and METTL14, can be related to poor prognosis of patients. This observation is consistent with the results of our study. Based on this result, we further explored the effects of METTL14 on the proliferation, migration, invasion and apoptosis of osteosarcoma. It is well known that an important factor affecting the prognosis of patients with osteosarcoma are early metastasis and recurrence after surgical treatment.4 To target the effect of METTL14 on tumor cell migration and invasion, we conducted a cell scratch test and transwell test. Consistent with what was expected, when the expression of METTL14 in cells is enhanced, it can inhibit the proliferation and migration of osteosarcoma cells. The cell clone formation experiment is an important technical method used to detect cell proliferation, invasiveness and sensitivity to killing factors. When a single cell proliferates in vitro for more than six generations, the cell population composed of its offspring becomes a colony or clone. Among them, the cell clone formation rate is the cell inoculation survival rate, which indicates the number of adherent cells that survived and formed clones after cell inoculation. The clone formation rate reflects the two important traits of cell population dependence and proliferation ability. In addition, cloning experiments can evaluate the tumorigenicity of cells in vivo. Cancer cells may not necessarily be tumorigenic in vivo. However, the stronger the cloning ability in vitro, the stronger the tumorigenicity in vivo. We carried out experiments on cell proliferation and cloning, and the results showed that METTL14 can inhibit cell viability and weaken cell cloning ability.
The activation of caspase-3 forms lysed caspase-3, and increased levels of lysed caspase-3 in tumor cells lead to apoptosis and secretion of paracrine factors.19 We tested the content of cleaved caspase-3 after overexpression of METTL14 and found a positive correlation between the two. We therefore hypothesized that METTL14 can increase the content of caspase-3 to promote osteosarcoma cell apoptosis. To verify this hypothesis, we first conducted a cell-level study. The results of PI staining and TUNEL staining showed that overexpression of METTL14 can promote tumor cell apoptosis. To further verify this hypothesis, we added the caspase inhibitor Z-VAD-FMK under the condition of overexpression of METTL14 to block the apoptosis caused by caspase-3 activation. The results showed a significant decrease in the number of apoptotic cells. It can be inferred from this that METTL14 induces tumor cell apoptosis after activating caspase-3. Although this finding has not definitively shown which particular pathway or gene (s) are affected by this activation, the specific mechanism can be studied through subsequent experiments. Our findings still lay the foundation for future research in understanding underlying mechanisms.
This study showed that METTL14 can promote osteosarcoma cell apoptosis, inhibit cell viability, and have a tumor suppressor effect on osteosarcoma. For any kind of cancer, early detection, early diagnosis and early treatment are the golden rules. As a type of early metastasis tumor, osteosarcoma should be diagnosed early and accurately. At present, the clinical diagnosis of this cancer is mainly based on the influence of the examination, but it is often difficult to accurately diagnose and predict osteosarcoma due to atypical manifestations of patients and numerous tumor subtypes. This study suggests that METTL14 has great potential as a clinical biomarker of osteosarcoma in the future and provides new ideas for the clinical treatment of osteosarcoma.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This work was supported by a grant from Postgraduate Research and Practice Innovation Program of Harbin Medical University (YJSKYCX2019-39HYD).
Disclosure
The authors indicated no potential conflicts of interest.
References
1. Simpson E, Brown HL. Understanding osteosarcomas. JAAPA. 2018;31(8):15–19. doi:10.1097/01.JAA.0000541477.24116.8d
2. Bielack SS, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20(3):776–790. doi:10.1200/JCO.2002.20.3.776
3. Wittig JC, et al. Osteosarcoma: a multidisciplinary approach to diagnosis and treatment. Am Fam Physician. 2002;65(6):1123–1132.
4. Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–5. doi:10.1093/annonc/mdq276
5. Kager L, Tamamyan G, Bielack S. Novel insights and therapeutic interventions for pediatric osteosarcoma. Future Oncol. 2017;13(4):357–368. doi:10.2217/fon-2016-0261
6. Zhang C, Fu J, Zhou Y. A review in research progress concerning m6a methylation and immunoregulation. Front Immunol. 2019;10:922. doi:10.3389/fimmu.2019.00922
7. Cao G, et al. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6(4):160003. doi:10.1098/rsob.160003
8. Liu ZX, et al. Link between m6a modification and cancers. Front Bioeng Biotechnol. 2018;6:89. doi:10.3389/fbioe.2018.00089
9. Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74(4):640–650. doi:10.1016/j.molcel.2019.04.025
10. Tong J, Flavell RA, Li HB. RNA m (6) A modification and its function in diseases. Front Med. 2018;12(4):481–489. doi:10.1007/s11684-018-0654-8
11. Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613. doi:10.1016/j.biopha.2019.108613
12. Ma S, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12(1):121. doi:10.1186/s13045-019-0805-7
13. He L, et al. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18(1):176. doi:10.1186/s12943-019-1109-9
14. Deng X, et al. RNA N (6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28(5):507–517. doi:10.1038/s41422-018-0034-6
15. Weng H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m (6) A modification. Cell Stem Cell. 2018;22(2):191–205 e9. doi:10.1016/j.stem.2017.11.016
16. Ma JZ, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N (6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–543.
17. Chen X, et al. METTL14 suppresses CRC progression via regulating N6-methyladenosine-dependent primary mir-375 processing. Mol Ther. 2020;28(2):599–612. doi:10.1016/j.ymthe.2019.11.016
18. Li J, et al. Dysregulated m6A-related regulators are associated with tumor metastasis and poor prognosis in osteosarcoma. Front Oncol. 2020;10:769. doi:10.3389/fonc.2020.00769
19. Shalini S, et al. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22(4):526–539. doi:10.1038/cdd.2014.216
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.