Back to Journals » Infection and Drug Resistance » Volume 15
Metagenomic Next-Generation Sequencing for Diagnostically Challenging Mucormycosis in Patients with Hematological Malignancies
Authors Zhang M, Lu W, Xie D, Wang J, Xiao X, Pu Y, Meng J, Lyu H, Zhao M
Received 21 October 2022
Accepted for publication 26 November 2022
Published 19 December 2022 Volume 2022:15 Pages 7509—7517
DOI https://doi.org/10.2147/IDR.S393201
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Meng Zhang,1,* Wenyi Lu,2,* Danni Xie,1 Jiaxi Wang,1 Xia Xiao,2 Yedi Pu,2 Juanxia Meng,2 Hairong Lyu,2 Mingfeng Zhao2
1First Center Clinic College of Tianjin Medical University, Tianjin, 300192, People’s Republic of China; 2Department of Hematology, Tianjin First Central Hospital, Tianjin, 300192, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Mingfeng Zhao; Hairong Lyu, Department of Hematology, Tianjin First Central Hospital, Tianjin, 300192, People’s Republic of China, Email [email protected]; [email protected]
Background: Metagenomic next-generation sequencing (mNGS) is a fast, sensitive and accurate diagnostic method for pathogens detection. However, reports on the application of mNGS in mucormycosis remain scarce.
Methods: From January 2019 to December 2021, we recruited 13 patients with hematological malignancies who were suspected of mucormycosis and completed mNGS in D20. Then we retrospectively analyze the clinical data, diagnosis, therapeutic process, and outcomes. In order to evaluate the diagnostic value of mNGS in hematological malignancies patients with suspected mucormycosis.
Results: All patients had high risk factors of Invasive Fungal Disease, including hematopoietic stem cell transplantation, immunosuppression, glucocorticoids, etc. The clinical presentations were pulmonary (n=9), rhino-orbito-cerebral (n=4). But the manifestations were nonspecific. All enrolled patients completed mNGS. And most (8/13, 61.54%) of the samples were from blood. Fungi can be detected in all specimens, including Rhizopus (n=7), Rhizomucor (n=4) and Mucor (n=2). In addition, 7/13 (53.85%) specimens were detected bacteria at the same time and virus were detected in 5/13 (38.46%). Histopathological examination was completed in 5 patients, 3 of which were completely consistent with the results of mNGS. After treatment, 6 patients were cured, while the other 7 patients died.
Conclusion: mNGS may be a complementary method for early diagnosis, especially for patients who are not suitable for histopathology examination or unable to obtain culture specimen. mNGS can also help detect bacteria and viruses simultaneously, allowing for appropriate and timely antibiotic administration and thus improving patient outcomes.
Keywords: metagenomic next-generation sequencing, mucormycosis, retrospectively, hematological malignancies, invasive fungal disease
Introduction
The rate of mucormycosis in patients with hematological malignancies are increasing year by year, especially those with immunosuppression, neutropenia, hematopoietic stem cell transplantation (HSCT) or other high-risk factors. Mucormycosis is a rare Invasive Fungal Disease (IFD) associated with high mortality. Studies have shown that delayed diagnosis and treatment increase the mortality of mucormycosis. If amphotericin B is delayed for ≥ 6 days, the mortality rate at 12 weeks will be doubled.1,2 Therefore, early diagnosis and treatment is the key to improve the outcome and reduce the mortality.
However, there was no characteristic signs and symptoms in patients with mucormycosis, and the diagnosis was difficult to determine in the early stage. At present, the diagnosis mainly depends on imaging examinations, histopathology and culture. Imaging changes such as multiple nodules (> 10) or pleural effusion may indicate mucormycosis, but the pathogenic diagnosis will be difficult because its characteristics are variable and similar to those of other invasive fungal infections.3,4 Histopathology or culture is the “gold standard” for diagnosing mucormycosis. Unfortunately, this is often difficult to be implemented in patients with hematologic malignancies because of severe thrombocytopenia.5 And positive culture result was obtained in only 50% of cases.6,7 Besides, other methods such as 1,3 beta-Dglucan detection test (G test) and Polymerase Chain Reaction (PCR) are also limited in the diagnosis of mucormycosis.8 Therefore, it is urgent to find new methods.
Metagenomic next-generation sequencing (mNGS) is a rapid microbial diagnosis method by analyzing the DNA / RNA content and abundance of microorganisms. It has the advantages of high sensitivity, short detection time, diagnose rare pathogen infections and so on.9 Lei Yang’s team has recommended biopsy-based or Bronchoalveolar Lavage Fluid -based mNGS for diagnosing pulmonary fungal infections.10 We further studied the diagnostic value of mNGS in patients with hematological malignancies and mucormycosis in this study.
Patients and Methods
Patients
Between January 2019 to December 2021, 13 patients with suspected mucormycosis in the Hematology Department of Tianjin First Central Hospital were enrolled in our study. The date when the patients suspected IFD was defined as D0. All enrolled patients completed mNGS within D20. According to the consensus of the European Research Group on cancer therapy and fungi (EORTC / MSG), 4 cases were finally “Proven” and 9 cases were “Probable”.11 The basic diseases, clinical manifestations, laboratory and imaging examinations when they were suspected, treatment and outcomes were analyzed retrospectively.
mNGS and Analyses
Samples were obtained from the peripheral blood or infection site, which was sent to BGI Laboratory within 12 hours after collection and nucleic acid was extracted. Using TIAN amp Micro DNA kit (DP316, Tiangen Biochemical Technology) kit and QIA amp Viral RNA Mini kit (52906 Qiagen) kit to extract DNA and RNA. Using SuperScript II reverse Transcription Kit (18064-014, Invitrogen) kit to reverse transcription of RNA into double-stranded complementary DNA (ds cDNA), which was ultrasonically broken into 200–300bp fragments. The sequence of the linker was circularized into a single-stranded circular structure. The circularized library was copied by a rolling circle to generate DNB nanospheres and loaded onto the sequencing chip. DNA libraries were constructed based on the Beijing Genomics Institute sequencer-100. After removing low-quality reads (< 35 bp) and computational subtracting human host sequences mapped to the human reference genome (hg19), high-quality sequences were generated.12 Microbial genome database was used to classify the remaining data, which were downloaded from NCBI (ftp://ftp.ncbi.nlm.nih.gov/genomes/). The pathogenic reading number refers to the number of strictly matched sequences of the microorganism detected at the genus/species level.
Diagnostic Criteria
Mucormycosis was defined based on the European Research Group on cancer therapy and fungi (EORTC / MSG).11 “Proven” patients need to meet culture and/or histopathological examination positive for mucormycosis. For “Probable” mucormycosis, imaging experts and clinicians of our hospital are required to make diagnosis together. Clinical remission refers to complete disappearance of the symptoms and signs, and obvious or complete absorption of the lesion site on imaging examination.
Results
Patients’ Characteristics
Of the 13 patients enrolled, 9 (69.23%) were males. The median age was 43 (14–73) years. The characteristics of those patients were listed in Table 1. Most patients were diagnosed with hematological malignancies, except one patient (ITP). All patients had high risk factors of IFD: HSCT or cell immunotherapy (n=7), immunosuppression or glucocorticoids (n=6), chemotherapy within 3 months (n=7), neutropenia (n=8), other high-risk factors (diabetes, malnutrition, etc).
 |
Table 1 Characteristics of Patients |
Pulmonary infection was considered in 9 patients, with others considered rhino-orbito-cerebral (n=4). And the clinical symptoms of patients are shown in Tables 2 and 3. Fever was the most common symptom (n=10), followed by headache (n=3). But these manifestations are nonspecific. The median time from symptom appearance to suspected IFD was 4 (1–19).
 |
Table 2 Characteristics of “Proven” Patients |
 |
Table 3 Characteristics of “Probable” Patients |
The mNGS Results
As shown in Figure 1, most (8/13, 61.54%) of our samples were from blood, others were from infection site (tissue, cerebrospinal fluid, swab). Fungi can be detected in all specimens, including Rhizopus (n=7), Rhizomucor (n=4) and Mucor (n=2).
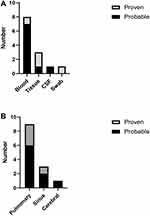 |
Figure 1 The distribution of mNGS sample and infection site. (A) Distribution of mNGS samples in the “Proven” and “Probable” group. (B) The infection site in the “Proven” and “Probable” group. |
In addition, 7/13 (53.85%) specimens were detected bacteria at the same time. The most frequently bacteria were enterococcus (n=3), followed by Haemophilus parainfluenzae (n=2), Pseudomonas aeruginosa (n=1) and Acinetobacter baumannii (n=1). Virus were detected in 5/13 (38.46%) specimens, including human herpesvirus type 5 (n=3), leptovirus (n=2) and human polyoma virus type 1 (n=2). Besides, bacteria, viruses and fungi were detected simultaneously in 2 specimens.
The Results of Traditional Detection Methods
Histopathological examination was completed in 5 patients (2 pulmonary tissue, 2 paranasal tissue and 1 sputum), 3 of which were completely consistent with the results of mNGS. In patient 3, mNGS detected Rhizomucor, while histopathology considered Mucor. 4 patients diagnosed “Proven” mucormycosis finally because one patient with negative results. It is worth noting that the results of mNGS in all patients were obtained before histopathology (Table 2).
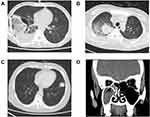
In 4 “Proven” patients, 3 patients were diagnosed pulmonary mucormycosis. Their chest CT mainly showed ground glass shadow, nodules and pleural effusion, which may indicate mucormycosis in previous reports (Figure 2). However, the result of sputum culture was Candida albicans in patient 1, and the other two cases were negative. Patient 4 was diagnosed paranasal mucormycosis, and his sinus computed tomography (CT) showed sinusitis. G test was negative in 4 “Proven” patients, and only patient 1 had positive Galactomannan test (GM) results.
In 9 “Probable” patients, 4 patients were not suitable for operation or biopsy due to coma, severe thrombocytopenia or critical condition. 6/9 patients were “Probable” pulmonary mucormycosis due to chest CT findings of nodules and pleural effusion. 2 head CT found intracranial infection, which may be caused by invasive fungus. Mucor was not found in the histopathology sinuses, but mNGS could detect it in patient 9. Combined with symptoms and other examinations, fungal infection cannot be completely excluded (Table 3).
The Influence of mNGS on Prognosis
On the basis of controlling the primary disease, 8 patients received amphotericin B combined with Posaconazole (1 received surgical debridement), 4 received amphotericin B treatment (1 received surgical debridement) and 1 did not receive treatment. In D90, 6/13 patients were cured. Bacteria were detected in 4/6 patients and viruses in 2/6 patients simultaneously. Unfortunately, 7/13 patients died, two of whom died of primary disease. Bacteria and virus were detected in 1/7 patient, bacteria in 2/7 patients, and virus in 1/7 patient simultaneously.
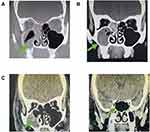
Patient 4 tested the mNGS of nasal swab at different times, and the result showed that the reading number of Rhizopus oryzae was 607 in D2. Combined with imaging examination and other examinations, the patient received amphotericin B combined with posaconazole, and received surgical debridement on D5. Rhizopus oryzae was detected in the nasal sinus tissue pathology, and the reading number of mNGS was decreased at D6 (467). The patient continued to receive antifungal treatment after surgery, and the reading of Rhizopus oryzae further decreased at D16 (48). At the same time, the patient’s nasal sinus CT also showed improvement (Figure 3). Therefore, mNGS may be useful in evaluating efficacy.
Discussion
Hematological malignancies are common basic diseases of mucormycosis,13 among which pulmonary infection is the most common.6,14 Consistent with this, more than half of the patients in our study considered pulmonary involvement. All of the included patients had multiple high-risk factors of mucormycosis, such as HSCT, agranulocytosis, immunosuppressive drugs, and so on. According to the clinical manifestations and imaging examinations of mucormycosis,3 these patients were highly suspected of mucormycosis, and finally 4 cases were finally “proven”. Because of the high mortality and disability rate of mucormycosis, early diagnosis is particularly important to improve the prognosis.
mNGS is a fast, sensitive and accurate diagnostic method, which has broad application prospects. It is less affected by the use of antibiotics and autoimmune status of patient.15 At present, many studies have explored the significance and value of mNGS in detection of bacterial, viral and fungal.16–19 Our previous research found that mNGS seems to be more meaningful for the diagnosis of fungal infection.20 Because mucormycosis is rare, the diagnostic value of mNGS is only reported in cases. It is preliminarily indicated that mNGS may be an effective tool for early diagnosis.21–25
We included 13 adult patients with hematological malignancies who were highly suspected of mucormycosis. mNGS results were all positive. And in 4 “Proven” patients, mNGS could get results before histopathology. For “Probable” patients, mNGS may help us find the pathogen, which is difficult for traditional methods. Although PCR can also lead to earlier diagnosis compared to culture, it requires prior knowledge of the suspected pathogen.16
As we describe, most of our samples are derived from peripheral blood, but that does not mean peripheral blood were infected sites. Because Mucor DNA fragments can easily enter the blood during a local infection.26 mNGS results need to be combined with clinical manifestations and images to locate the sites of infection. In our research, 4 patients were not suitable for biopsy due to thrombocytopenia or coagulation dysfunction. Their peripheral blood mNGS all suggested mucormycosis, then combined with traditional methods and clinical manifestations, the site of infection was identified. This combination of methods may be more important to those patients.
Qingya Cui reported a case of acute T-lymphocyte leukemia, several pathogens were detected by using mNGS. In particular, mNGS identified Malassezia pachydermum, Mucor racemosus, and Lauteria mirabilis in the peripheral blood and local secretion samples. The Mucor and bacterial infections were further confirmed via multi-site and repeated fungal and bacterial cultures, respectively.25 Patients with immunodeficiency may be infected with bacteria, viruses and fungi at the same time. Early detection of mixed infection is also important. mNGS can detect bacteria, fungi and viruses simultaneously. In our study, 53.85% of patients detected bacteria at the same time, and 38.46% detected virus. It seems that appropriate and timely antibiotic administration according to the results of mNGS seems to better control the symptoms. In addition, we found that the pathogenic reading number seems to help us evaluate the progress of the disease. It may help us to adjust the use of antifungal timely.
mNGS combined with traditional detection methods (imaging examinations, histopathology and culture) can make the disease quickly diagnosed. Therefore we believe that mNGS may be a complementary method for early diagnosis, especially for patients who are not suitable for histopathology or culture. mNGS can also help detect bacteria and viruses simultaneously, allowing appropriate and timely use of antibiotics, thereby improving the prognosis of patients.
Ethics Statement
The study was approved by the Ethics Committee of Tianjin First Central Hospital. And our study complies with the Declaration of Helsinki. The need for informed consent was waived due to the retrospective nature of the study and because the data were anonymously analyzed. Written informed consent was obtained from the individual for the publication of any potentially identifiable images or data included in this article.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by grants from the General Project of National Natural Science Foundation of China (81970180 to MZ), and the Key Science and Technology Support Project of Tianjin Science and Technology Bureau (20YFZCSY00800 to MZ), as well as Tianjin Key Medical Discipline (Specialty) Construction Project (TJYXZDXK-056B).
Disclosure
The authors report the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Chamilos G, Lewis RE, Kontoyiannis DP. Delaying amphotericin B-based frontline therapy significantly increases mortality among patients with hematologic malignancy who have zygomycosis. Clin Infect Dis. 2008;47(4):503–509. doi:10.1086/590004
2. Jeong SJ, Lee JU, Song YG, Lee KH, Lee MJ. Delaying diagnostic procedure significantly increases mortality in patients with invasive mucormycosis. Mycoses. 2015;58(12):746–752. doi:10.1111/myc.12428
3. Chamilos G, Marom EM, Lewis RE, Lionakis MS, Kontoyiannis DP. Predictors of pulmonary zygomycosis versus invasive pulmonary aspergillosis in patients with cancer. Clin Infect Dis. 2005;41(1):60–66. doi:10.1086/430710
4. Lass-Florl C, Resch G, Nachbaur D, et al. The value of computed tomography-guided percutaneous lung biopsy for diagnosis of invasive fungal infection in immunocompromised patients. Clin Infect Dis. 2007;45(7):e101–4. doi:10.1086/521245
5. Skiada A, Lanternier F, Groll AH, et al. Diagnosis and treatment of mucormycosis in patients with hematological malignancies: guidelines from the 3rd European Conference on Infections in Leukemia (ECIL 3). Haematologica. 2013;98(4):492–504. doi:10.3324/haematol.2012.065110
6. Roden MM, Zaoutis TE, Buchanan WL, et al. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin Infect Dis. 2005;41(5):634–653. doi:10.1086/432579
7. Walsh TJ, Gamaletsou MN, McGinnis MR, Hayden RT, Kontoyiannis DP. Early clinical and laboratory diagnosis of invasive pulmonary, extrapulmonary, and disseminated mucormycosis (zygomycosis). Clin Infect Dis. 2012;54(Suppl 1):S55–60. doi:10.1093/cid/cir868
8. Bellanger AP, Grenouillet F, Henon T, et al. Retrospective assessment of beta-D-(1,3)-glucan for presumptive diagnosis of fungal infections. APMIS. 2011;119(4–5):280–286. doi:10.1111/j.1600-0463.2011.02728.x
9. Grumaz S, Stevens P, Grumaz C, et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016;8(1):73. doi:10.1186/s13073-016-0326-8
10. Yang L, Song J, Wang Y, Feng J. Metagenomic next-generation sequencing for pulmonary fungal infection diagnosis: lung biopsy versus bronchoalveolar lavage fluid. Infect Drug Resist. 2021;14:4333–4359. doi:10.2147/IDR.S333818
11. Donnelly JP, Chen SC, Kauffman CA, et al. Revision and update of the consensus definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin Infect Dis. 2020;71(6):1367–1376. doi:10.1093/cid/ciz1008
12. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi:10.1093/bioinformatics/btp324
13. Skiada A, Pavleas I, Drogari-Apiranthitou M. Epidemiology and diagnosis of mucormycosis: an update. J Fungi. 2020;6(4):265. doi:10.3390/jof6040265
14. Tedder M, Spratt J, Anstadt M, Hegde S, Tedder S, Lowe J. Pulmonary mucormycosis: results of medical and surgical therapy. Ann Thorac Surg. 1994;57(4):1044–1050. doi:10.1016/0003-4975(94)90243-7
15. Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67(suppl_2):S231–S40. doi:10.1093/cid/ciy693
16. Gu W, Deng X, Lee M, et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med. 2021;27(1):115–124. doi:10.1038/s41591-020-1105-z
17. Besser J, Carleton HA, Gerner-Smidt P, Lindsey RL, Trees E. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin Microbiol Infect. 2018;24(4):335–341. doi:10.1016/j.cmi.2017.10.013
18. Zanella MC, Cordey S, Laubscher F, et al. Unmasking viral sequences by metagenomic next-generation sequencing in adult human blood samples during steroid-refractory/dependent graft-versus-host disease. Microbiome. 2021;9(1):28. doi:10.1186/s40168-020-00953-3
19. Wang C, You Z, Fu J, et al. Application of metagenomic next-generation sequencing in the diagnosis of pulmonary invasive fungal disease. Front Cell Infect Microbiol. 2022;12:949505. doi:10.3389/fcimb.2022.949505
20. Zhang M, Wang Z, Wang J, et al. The value of metagenomic next-generation sequencing in hematological malignancy patients with febrile neutropenia after empiric antibiotic treatment failure. Infect Drug Resist. 2022;15:3549–3559. doi:10.2147/IDR.S364525
21. Liu Y, Zhang J, Han B, et al. Case report: diagnostic value of metagenomics next generation sequencing in intracranial infection caused by mucor. Front Med. 2021;8:682758. doi:10.3389/fmed.2021.682758
22. Sun Y, Li H, Chen J, et al. Case report: metagenomics next-generation sequencing can be performed for the diagnosis of disseminated mucormycosis. Front Med. 2021;8:675030. doi:10.3389/fmed.2021.675030
23. Wen B, Cai L, Cai Y, Du X. Case report: metagenomics next-generation sequencing for diagnosing cerebral infarction and infection caused by hematogenous disseminated mucormycosis in a patient with acute lymphoblastic leukemia. Front Med. 2021;8:779981. doi:10.3389/fmed.2021.779981
24. Chen L, Su Y, Xiong XZ. Rhizopus microsporus lung infection in an immunocompetent patient successfully treated with amphotericin B: a case report. World J Clin Cases. 2021;9(35):11108–11114. doi:10.12998/wjcc.v9.i35.11108
25. Zhang J, Luo J, Weng X, et al. A case report of the metagenomics next-generation sequencing for early detection of central nervous system mucormycosis with successful rescue in patient with recurrent chronic lymphocytic leukemia. Ann Transl Med. 2022;10(12):722. doi:10.21037/atm-22-2533
26. Li N, Cai Q, Miao Q, Song Z, Fang Y, Hu B. High-throughput metagenomics for identification of pathogens in the clinical settings. Small Methods. 2021;5(1):2000792. doi:10.1002/smtd.202000792
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.