Back to Journals » Breast Cancer: Targets and Therapy » Volume 9
Mentha arvensis (Linn.)-mediated green silver nanoparticles trigger caspase 9-dependent cell death in MCF7 and MDA-MB-231 cells
Authors Banerjee PP , Bandyopadhyay A , Harsha SN, Policegoudra RS, Bhattacharya S, Karak N , Chattopadhyay A
Received 22 December 2016
Accepted for publication 9 March 2017
Published 18 April 2017 Volume 2017:9 Pages 265—278
DOI https://doi.org/10.2147/BCTT.S130952
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Pranela Rameshwar
Prajna Paramita Banerjee,1 Arindam Bandyopadhyay,1 Singapura Nagesh Harsha,2 Rudragoud S Policegoudra,3 Shelley Bhattacharya,4 Niranjan Karak,2 Ansuman Chattopadhyay1
1Molecular Genetics Laboratory, Department of Zoology, Visva-Bharati, Santiniketan, West Bengal, 2Advanced Polymer and Nanomaterial Laboratory, Department of Chemical Sciences, Center for Polymer Science and Technology, Tezpur University, Napaam, 3Division of Pharmaceutical Technology, Defence Research Laboratory, Tezpur, Assam, 4Environmental Toxicology Laboratory, Department of Zoology, Visva-Bharati, Santiniketan, West Bengal, India
Introduction: Leaf extract of Mentha arvensis or mint plant was used as reducing agent for the synthesis of green silver nanoparticles (GSNPs) as a cost-effective, eco-friendly process compared to that of chemical synthesis. The existence of nanoparticles was characterized by ultraviolet–visible spectrophotometry, dynamic light scattering, Fourier transform infrared spectroscopy, X-ray diffraction, energy-dispersive X-ray analysis, atomic-force microscopy and transmission electron microscopy analyses, which ascertained the formation of spherical GSNPs with a size range of 3–9 nm. Anticancer activities against breast cancer cell lines (MCF7 and MDA-MB-231) were studied and compared with those of chemically synthesized (sodium borohydride [NaBH4]-mediated) silver nanoparticles (CSNPs).
Materials and methods: Cell survival of nanoparticle-treated and untreated cells was studied by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay. Cell-cycle analyses were carried out using fluorescence-activated cell sorting. Cell morphology was observed by fluorescence microscopy. Expression patterns of PARP1, P53, P21, Bcl2, Bax and cleaved caspase 9 as well as caspase 3 proteins in treated and untreated MCF7 and MDA-MB-231 cells were studied by Western blot method.
Results: MTT assay results showed that Mentha arvensis-mediated GSNPs exhibited significant cytotoxicity toward breast cancer cells (MCF7 and MDA-MB-231), which were at par with that of CSNPs. Cell cycle analyses of MCF7 cells revealed a significant increase in sub-G1 cell population, indicating cytotoxicity of GSNPs. On the other hand, human peripheral blood lymphocytes showed significantly less cytotoxicity compared with MCF7 and MDA-MB-231 cells when treated with the same dose. Expression patterns of proteins suggested that GSNPs triggered caspase 9-dependent cell death in both cell lines. The Ames test showed that GSNPs were nonmutagenic in nature.
Conclusion: GSNPs synthesized using Mentha arvensis may be considered as a promising anticancer agent in breast cancer therapy. They are less toxic and nonmutagenic and mediate caspase 9-dependent apoptosis in MCF7 and MDA-MB-231 cells.
Keywords: nanoparticles, EDX, TEM, breast cancer cells, anticancer, nonmutagenic
Introduction
The field of nanotechnology is one of the most potential areas of research in modern medical science. Design, manipulation and application of materials with a size range of 1–100 nm have gained a reasonable interest in recent times due to their unique biological and medicinal applications. Nanoparticles exhibit absolutely novel or enhanced properties based on specific characteristics such as size, distribution and morphology compared with larger particles of the bulk materials from which they are made.1 There is an increasing quest to develop high-yield, low-cost, nontoxic and environment-friendly nanoparticles.
The potentials and promises of plant system in the biologically assisted synthesis of metal nanoparticles called green synthesis have now become a key issue in nanoscience research.2 These methods use biological compounds such as microorganisms,3 enzymes,4 fungi,5 amino acids6 and plants or plant extracts.7 The use of biocompounds as reducing or stabilizing agents can be a possible eco-friendly alternative to chemical and physical methods.
Silver is known as an efficient antimicrobial agent showing lower toxicity to mammalian cells.8 Physicochemical properties of silver nanoparticles (SNPs) play a significant role in the field of biology and medicine. Reports are available regarding the synthesis of SNPs using various plant sources such as lingonberry and cranberry juices,9 Pistacia atlantica seeds10 and Thuja occidentalis leaf extract.7
According to the latest world cancer statistics, an estimated number of 14.1 million new cancer cases and 8.2 million cancer-related deaths occurred in 2014.11 GLOBOCAN 2012 statistics also reported >20% increase in breast cancer incidence and 14% increase in mortality. Breast cancer has become the most common cause of cancer death among women (522,000 deaths in 2012) and the most frequently diagnosed cancer among women worldwide. In the less developed countries, it is the leading cause of cancer death.12 The incidence rate of breast cancer is higher in developing, middle-income and low-income countries.13,14
SNPs synthesized using different plant extracts showed a significant cytotoxic effect on MCF7 cells,7,15,16 HeLa cells,7,17,18 NIH3T3 fibroblast cells,19 human glioblastoma cells20 and HEp2 cells.21 Further studies in this direction are essential to search for SNPs that have higher toxicity against cancer cells but lesser toxicity toward normal cells. In the present study, we analyzed the anticancer potential of SNPs synthesized from the leaves of Mentha arvensis (green silver nanoparticles [GSNPs]), commonly known as wild/field mint or pudina, a tropical ethnomedicinal plant, in MCF7 and MDA-MB-231 breast cancer cell lines, and the same was compared with that of chemically synthesized (sodium borohydride [NaBH4]-mediated) silver nanoparticles (CSNPs). Their mutagenic properties were also evaluated in the present study.
Materials and methods
Materials
Mentha arvensis leaves were collected from the state of Assam, India. Silver nitrate (AgNO3), poly(ethylene glycol) (PEG) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitro blue tetrazolium (NBT) were purchased from Merck (India). Dulbecco’s Modified Eagle’s Medium (DMEM), Roswell Park Memorial Institute (RPMI)-1640 culture media, fetal bovine serum (FBS), penicillin–streptomycin antibiotic solution, phytohemagglutinin (PHA), 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) and radioimmunoprecipitation assay (RIPA) buffer were purchased from HiMedia (Mumbai, India). Heparin, ethidium bromide (EtBr) and trypan blue were purchased from SRL (India); poly-l-lysine, Histopaque, bisbenzimide (Hoechst 33342), paraformaldehyde, acridine orange, propidium iodide (PI), RNase, NaBH4 and anti-human primary antibodies used were procured from Sigma-Aldrich (St Louis, MO, USA). ALP-linked goat anti-rabbit secondary antibodies were purchased from Abcam (UK).
Preparation of nanoparticles
Approximately 2.0 g of Mentha arvensis leaves were washed with tap water to remove soil particles and further with de-ionized water and homogenized using a domestic blender. The aqueous extract was prepared by stirring for ~20 min in 50 mL of water at 50°C, followed by filtration through a muslin cloth. A total of 2 mL of the extract was added to 0.01 M AgNO3 in 25 mL of 5% (w/v) PEG solution. The formation of GSNPs was indicated by a gradual change (brown) in color.
Characterization of nanoparticles
The optical property of GSNPs was analyzed by ultraviolet–visible (UV–Vis) spectroscopic studies at room temperature, operated at a resolution of 1 nm between 200 and 800 nm ranges (Hitachi U-2001; Hitachi, Tokyo, Japan). The size distribution of GSNPs was measured using dynamic light scattering (DLS; Zetasizer Nano ZS ZEN3600; Malvern Instruments, Malvern, UK). Fourier transform infrared spectroscopy (FTIR) spectra of the GSNPs were recorded using a Nicolet (Madison, WI, USA) FTIR Impact 410 spectrophotometer using KBr pellets. An X-ray diffractometer, “MiniFlex” (Rigaku Corporation, Japan), was used for the analysis of GSNPs sample at room temperature (24°C) at the scanning rate of 2.0°min−1 over the range of 2θ = 10–80°. Elemental compositions of the synthesized GSNPs were examined through energy-dispersive X-ray analysis (EDX) by using the JOEL scanning electron microscope (SEM). The sample was spin coated onto a freshly peeled off mica sheet (which is hydrophilic) and used for atomic-force microscopy (AFM) studies. AFM analysis was done using MultiMode SPM (Digital Instruments, Santa Barbara, CA, USA) equipped with NanoScope IV A controller. Force modulation etched silicon probes (Veeco) tips of force constant 1−5 N/m and the resonance frequency of 75 kHz were used for imaging the sample; measurements were done in the tapping mode, and the images were analyzed using the software (NanoScope) provided with the instrument. Transmission electron microscopy (TEM) analysis was carried out to analyze the size and morphology of synthesized nanoparticles (Tecnai G2 ST LaB6; FEI, USA) in which the samples were prepared by drop coating the GSNPs solution onto the carbon-coated copper grid.
Cell culture
MCF7 and MDA-MB-231 cell lines were purchased from the National Centre for Cell Science (Pune, India). Cell culture was done according to the description of Barua et al.7 Cells were grown in DMEM, supplemented with 10% (v/v) FBS and 1% penicillin–streptomycin solution and incubated at 37°C in a humidified 5% CO2 incubator (Thermo Fisher Scientific, Waltham, MA, USA).
Cell viability assay
Cells (1 ¥ 104) were seeded into each well of a 96-well plate and allowed to stand for 24 h. After that, complete media were removed and the cells were replenished with incomplete media (without FBS) and treated with different concentrations of SNPs. Both negative and positive controls were used along with the treated groups. Cells were further incubated at 37°C in the presence of 5% humidified CO2 for 48 h, and the proliferation rates were estimated by the MTT assay at 595 nm using a Beckman Coulter spectrophotometer (USA). The percentage of viable cells was calculated taking viability of untreated cells as 100%.
Cell morphology study
Approximately 4 × 104 cells (MCF7, MDA-MB-231) were plated in each well of a six-well plate and incubated in a humidified 5% CO2 incubator at 37°C until cells reached 70% confluency and were treated with 1.56 and 12.5 µg/mL of GSNPs. Alteration in the cell morphology was studied under an inverted phase contrast microscope (Dewinter, Italy), and images were captured at different time points using the Biowizard software.
Hoechst staining
Approximately 104 cells were taken from the 96-well plate after treatment with GSNPs, washed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde. Cells were placed on poly-l-lysine-coated slides, dried in air, stained with Hoechst for 10 min and washed with PBS. Stained cells were examined under a fluorescent microscope (Dewinter) and analyzed for nuclear fragmentation.
Acridine orange (AO)/ethidium bromide (EtBr) staining
MCF7 and MDA-MB-231 cells were plated in six-well plates (4 × 104 cells/well) separately and incubated in the humidified 5% CO2 incubator at 37°C until cells reached 70% confluency. Cells were treated with varying concentrations of GSNPs, incubated for 48 h and harvested and stained with AO/EtBr dye mix (one part of 100 µg/mL of AO and one part of 100 µg/mL of EtBr in PBS) on a clean grease-free microscope slide. Live, apoptotic and necrotic cells were observed under the fluorescent microscope at a magnification of 100¥. Experiments were repeated three times.
Flow cytometric analysis
After 48 h of treatment, MCF7 cells were fixed in ethanol, washed with PBS and resuspended in 500 µL of PI solution (50 µg/mL PI, 0.2 mg/mL RNase). The cell suspension was incubated for 30 min at room temperature in the dark. From each sample, a large number of cells (10,000) were analyzed in FACSCalibur (Becton Dickinson, USA) instrument, and the data were interpreted using CellQuest Pro software to estimate the number of cells distributed in different phases of the cell cycle.
Western blot analysis
MCF7 and MDA-MB-231 cells were collected, washed with ice-cold PBS and lysed in chilled RIPA buffer. The supernatants were collected after centrifugation at 12,000 rpm at 4°C for 30 min. Protein content was quantified using Lowry et al’s method.22 An equal amount of protein samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) at a constant voltage (60 V) for 2.5 h and then electroblotted onto polyvinylidene fluoride (PVDF) membranes using a blotting apparatus (Bio-Rad Laboratories Inc., Hercules, CA). The membranes were blocked (1 h, room temperature) in powdered skim milk (5%; HiMedia) in Tris-buffered saline–Tween 20 (TBST; pH 8.0) and subsequently incubated (1 h, room temperature) with primary antibodies at 4°C overnight, followed by 2 h incubation with the corresponding ALP-linked goat anti-rabbit (for PARP1, Bcl2, cleaved caspase 9 and caspase 3) or anti-mouse (for P53, P21, Bax and β-actin) secondary antibodies with continuous rocking. The immunoreactive bands were detected using BCIP/NBT. Densitometric quantification was done by ImageJ (NIH) software.
Cell viability assay of human peripheral blood lymphocytes (HPBLs)
Fresh blood was collected from five healthy nonsmokers, non-alcoholic male donors (21–25 years of age) with written informed consents by venipuncture and stored into heparinized falcon tubes. Blood was collected by pathologists employed at the university hospital under the supervision of medical doctors. All studies were performed complying fully with the approved “Ethical Guidelines for Biomedical Research on Human Subjects” formulated by the Indian Council of Medical Research, India. The work was reviewed and approved by the Institutional Ethics Committee for Human Research of Visva-Bharati University.
Lymphocytes (HPBLs) were isolated according to the method of Bøyum.23 Blood was diluted with an equal volume of PBS (Ca2+, Mg2+ free; pH 7.4) and layered over 2 mL of Histopaque and centrifuged at 1000 × g for 30 min. The buffy coat was aspirated into 3–5 mL PBS and centrifuged at 1000 × g for 10 min, and the washing process was repeated thrice. The pellet was resuspended in RPMI-1640 media (1 × 106 cells/mL), and viability was checked using the trypan blue dye exclusion method.24 Cell viability was found to be >95%, which indicated successful isolation.
Isolated HPBLs (0.5 mL) were stimulated by PHA and cultured in 5 mL of RPMI-1640 media with 10% FBS along with antibiotics. After 24 h, 12.5 µg/mL of nanoparticles (GSNPs and CSNPs) were added to triplicate samples and cultured for another 48 h. Both negative and positive controls were used along with the treated groups. After 48 h, MTT was added and cell viability was checked at 595 nm using a Beckman Coulter spectrophotometer. Percentage of viable cells was calculated taking the viability of untreated cells as 100%.
Mutagenicity test
The assay was performed according to the plate incorporation procedure described by the Organization for Economic Co-operation and Development (OECD) Test Guideline 471 recommendations25 and Maron and Ames.26 The tester strains of Salmonella typhimurium (TA98, TA100, TA1535 and TA1538) used in this study were purchased from the Institute of Microbial Technology (IMTECH), Chandigarh, India. Overnight (16–18 h) bacterial cultures in nutrient broth (HiMedia) were used for the assay. Approximately 0.1 mL of freshly grown overnight bacterial culture was mixed with different concentrations (0.6, 1.2, 2.5 and 5 mg/plate) of test samples and 2 mL of top agar (containing 0.5% NaCl and 0.6% agar) supplemented with 0.5 mM L-histidine and d-biotin. The entire mixture was gently poured on the minimal glucose agar plates. Plates were incubated for 48–72 h at 37°C. The mutagenicity of the test samples was expressed by counting revertant bacterial colonies (His+). Experiments with all the strains were conducted with and without the addition of sodium phosphate buffer (0.1 mM, pH 7.4). Negative and positive controls were used simultaneously in each experiment. The positive controls used in the assays performed without metabolic activation system were sodium azide (SA) for TA100 and TA1535 and 4-Nitro-o-phenylenediamine (4-NPD) for TA98 and TA1538 strains. Triplicates were maintained for each test concentration against all bacteria strains.
Statistical analysis
The results were expressed as mean ± standard error (SE). Statistical analysis of the data was performed by paired t-test, and p<0.05 were considered significant.
Results
Preparation and characterization of nanoparticles
UV–Vis spectroscopy
The colorless AgNO3 solution turned brown (Figure 1), indicating the formation of GSNPs. The AgNO3 solution was colorless, and the Mentha arvensis leaf extract was green before the start of the reaction. The reaction mixture became yellow-brown within 2 h when treated with the Mentha arvensis leaf extract at room temperature, showing a preliminary indication of the formation of GSNPs. Figure 2 shows the UV–Vis spectra recorded for the GSNPs. It was observed that the absorbance peak appeared at ~436 nm. For spherical GSNPs, the characteristic surface plasmon resonance (SPR) absorption band of GSNPs was observed in the range of 400–475 nm.27 The nanoparticles were kept at room temperature for stability test. They were found to be stable for at least 6 months from the day of preparation. However, for all experimental purposes, only freshly prepared nanoparticles were used.
DLS measurement
DLS was used to measure the particle size in the colloidal solution. The presence of bio-organics in the plant extract will cause the size of particles to increase. It could also be due to the various forces of interaction in the solution such as the Van der Waals force of attraction. The DLS-measured size will be slightly bigger because the DLS method measures the hydrodynamic radius.28 The DLS pattern revealed that GSNPs, synthesized by this method, had a zeta average of 145.2 nm and polydispersity index (PDI) of 0.226 (Figure 3).
FTIR measurement
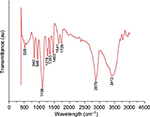
The FTIR spectroscopy is used to identify a major compound that is responsible for the biological reduction of silver ions (Ag+) into SNPs (Agº) from the leaf extract of Mentha arvensis. The major bands in the FTIR spectrum of GSNPs were observed at 3412, 2878, 1463, 1353, 1278 and 1106 cm-1; the minor bands were observed at 1729, 1641, 945, 840 and 526 cm-1 (Figure 4). A distinct peak of GSNPs seen at 3412 cm-1 was due to the O–H stretching vibrational frequencies, and bands in these regions strongly indicate the presence of an organic molecule with an alcohol functional group. A significant band at a stretching frequency of 2878 cm-1, which indicates the alkyl C–H group, suggests the presence of saturated molecules. A distinct band at 1641 cm-1 was assigned to the C=C stretching vibrations in the amide linkages of proteins.29 In particular, the 1278 cm-1 arises most probably from the C–O group of polyols such as hydroxyl flavones.30 The band at 1353 cm-1 with characteristic C-N stretch represents an aliphatic amine group and bands at 1463 cm-1 and 1729 cm–1 indicate the presence of C-H bending and C=O, carbonyl group respectively. The bands at 576 and 526 cm-1 correspond to C–X (alkyl halides). It is well known that bioconstituents interact with silver ion salts and mediate the reduction process with functional groups.
X-ray diffraction (XRD) measurement
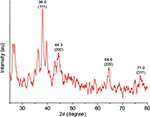
An XRD spectrum (Figure 5) clearly illustrates the crystalline nature of the synthesized GSNPs. The XRD peaks at 2θ degrees of 38.0, 44.3, 64.6 and 77.0 can be ascribed to the (111), (200), (220) and (311) crystalline planes of face-enhanced cubic (fcc) crystalline structure of metallic nanoparticles.31 Some additional unassigned peaks were also observed in the XRD study due to the presence of bioorganic matters and capping agents for GSNP formation.32
  | Figure 5 XRD spectra of Mentha arvensis leaf extract-mediated SNPs. Abbreviations: XRD, X-ray diffraction; SNP, silver nanoparticle; au, atomic unit. |
EDX measurement
EDX provides information on the chemical composition of the specific locations. Figure 6A shows the SEM image from which the EDX spectra analysis was carried out, showing a representative profile of the EDX analysis obtained by focusing on GSNPs (Figure 6B). From Figure 6C–E, the elemental mapping was carried out, which showed the presence of carbon, oxygen, and silver in the synthesized GSNPs.
AFM measurement
AFM analysis of the synthesized GSNPs was made, and the AFM image is shown in Figure 7, which confirms the spherical shape of the particles. GSNPs of size ~2.8–9.9 nm were obtained using the Mentha arvensis extract.
TEM measurement
A transmission electron microscope was used to analyze the size and shape of the formed nanoparticles. The TEM image of GSNPs illustrates that they were spherical in shape with an average size of ~4–9 nm (Figure 8).
Viability of MCF7 and MDA-MB-231 cells
Cell viability was measured by the MTT assay using different concentrations of SNPs (Figure 9). The results showed that cell viability decreased with the increasing concentration of the nanoparticles. In both the breast cancer (MCF7 and MDA-MB-231) cell lines, >50% cell death occurred at a concentration of 6.25 µg/mL of GSNPs and <30% of the cells survived after treatment with 12.5 µg/mL GSNPs, which was even higher than that of mitomycin-treated cells. The assay confirmed that the prepared nanoparticles were effective against human breast cancer cell lines, although the effect was most pronounced in MCF7 cell lines followed by MDA-MB-231. Cell viability assay with CSNPs revealed that the GSNPs were equally effective compared with same doses of CSNPs (Figure 9).
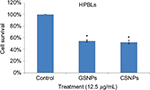
Cell viability of HPBLs
Cell viability of HPBLs (Figure 10) was assessed to perceive the toxicity of synthesized GSNPs toward normal cells. GSNP (12.5 µg/mL)-treated HPBLs showed 54.5% cell viability after 48 h compared to the untreated cells, the viability of which was taken as 100%.
Therefore, the GSNPs were found to be active against both cancer and normal cells, although the cytotoxicity toward normal cell was not significant.
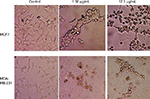
Cell morphology
Changes in cell morphology were observed after exposure of breast cancer cells (MCF7 and MDA-MB-231) to GSNPs (1.56 and 12.5 µg/mL; Figure 11). After 48 h of exposure, normal cellular morphology was distorted significantly in both cases compared to the control. Exposure to GSNPs reduced both cell size and number, and no recovery was observed until 96 h. Cells lost their typical morphology and adhesion property and were clumped on treatment with GSNPs. From the microscopic study, it was abundantly clear that synthesized GSNPs treatment can induce death and severe morphological alterations in human breast cancer cells.
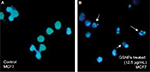
Nuclear fragmentation study by Hoechst staining
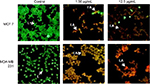
GSNP-induced nuclear fragmentation was studied through Hoechst staining (Figure 12). Nuclear condensation and fragmentation were evident and prominent in GSNP-treated cancer cells. In the control group, the cells showed intact nuclear architecture. This indicated that one of the possible causes of cell death that occurred in the GSNP-treated cancer cells was due to DNA damage and the inability of the cells to repair the damage. The extensive nuclear fragmentations might have also activated the apoptotic pathways.
AO/EtBr staining
In breast cancer cells, the type of cell death induced by GSNPs was monitored under the fluorescent microscope. Live cells with normal morphology were seen in the control group. Both live and early apoptotic cells were seen in the presence of 1.56 µg/mL GSNPs, while in the presence of 12.5 µg/mL GSNPs, nearly all were late apoptotic cells (Figure 13). Untreated MCF7 and MDA-MB-231 cells showed 94% and 95.6% live cells, respectively. When treated with 1.56 µg/mL GSNPs, MCF7 and MDA-MB-231 cells exhibited 74% and 80% early apoptotic cells. Treatment with 12.5 µg/mL GSNPs showed 30% early apoptotic cells with 65% late apoptotic cells in MCF7 and 10% early apoptotic cells with 87% late apoptotic cells in case of MDA-MB-231.
Morphological changes of apoptosis, including cell shrinkage and chromatin condensation, compared to control cells (Figure 12) suggest that treatment with GSNPs significantly increased apoptosis in MCF7 and MDA-MB-231 cells in a dose-dependent manner.
Flow cytometric analysis
The fluorescence-activated cell sorting (FACS) analysis of MCF7 cells after treatment with 12.5 µg/mL GSNPs showed a significant increase in the fraction of cells present in the sub-G1 phase and subsequent decline of cells at the G1 phase compared to untreated control cells (Figure 14), which might be due to apoptosis.
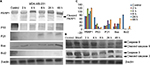
Western blot analysis
Expression patterns of PARP1, P53, P21, Bax, Bcl2, cleaved caspase 9 and caspase 3 were observed in controlled and treated cells at various time points (2, 4, 8, 24 and 48 h; Figures 15 and 16). Upregulation of PARP1, P53, P21, Bax and cleaved caspase 9 was observed in MCF7 cells after GSNPs treatment, whereas Bcl2 was downregulated. Cleaved bands of PARP1 were prominent at 24 and 48 h after treatment with GSNPs. In MDA-MB-231 cells, the mutant P53 protein was downregulated, whereas PARP1, P53, P21, Bax, cleaved caspase 9 and both procaspase 3 and cleaved caspase 3 proteins were upregulated. Cleavage of PARP1 protein was noticed 2 h onward, which was prominent at 8 and 48 h.
Mutagenicity analysis
The number of revertant colonies observed in each plate was less than two times the number of revertant colonies of the spontaneous mutation, and no dose-dependent response was exhibited by the GSNPs up to 5 mg per plate concentration (Table 1). The mutagenicity test by the Ames test revealed that the GSNPs have no mutagenic potential toward all the four strains tested.
Discussion
The most common type of cancer in the US is breast cancer, with >249,000 new cases expected in 2016 as per the cancer incidence and mortality statistics reported by the American Cancer Society.33 In underdeveloped and developing countries such as India, the incidence of breast cancer is also increasing alarmingly, and due to lack of awareness and infrastructural facility, the cancer cases are not detected at the early stages. Extremely high cost of chemotherapy is a major obstacle to continue and complete the course of treatment, resulting in a high mortality rate among patients. Not only patients lose their lives, but the whole family of the deceased also suffers from a huge financial burden to meet the expenses of chemotherapy. Therefore, cancer cases in such countries not only are a medical issue but should also be considered as a factor leading to the major socioeconomic crisis. This high-cost treatment of cancer patients needs immediate attention to look for alternative medicines in developing and underdeveloped countries. Therefore, it is important to find cost-effective, environment-friendly alternative approaches to synthesize SNPs with potential anticancer attributes.
In this regard, biosynthesis of nanoparticles has received considerable attention as an environment-friendly technology in material sciences. A great importance has been given to the biosynthesis of metal nanoparticles using plants.34 To the best of our knowledge, this is the first report on the use of Mentha arvensis leaves for the synthesis of SNPs. The qualitative phytochemical analysis of Mentha arvensis showed the presence of vital secondary metabolites, such as flavonoids, phenols and saponins.
Characterizations of GSNPs showed that the Mentha arvensis leaf extract can be used as an effective reducing agent for the synthesis of nanoparticles, which act as a potent anticancer agent against MCF7 and MDA-MB-231 cell lines. The level of cytotoxicity at equivalent doses was comparable to that of CSNPs and was significantly less in normal HPBLs. This difference might be attributed to the leaky vasculature of the rapidly growing cancer cells, which allows the nanoparticles to penetrate easily through the membrane and exert their effect.
This study showed that estrogen receptor-positive MCF7 cells were more sensitive to a lower dose (1.56 µg/mL) than the MDA-MB-231 cells (estrogen receptor negative), but at higher doses, the effects were reversed. Inhibition of cell growth could be due to either inhibition of cell proliferation or death of cells. Flow cytometric analysis revealed an increase of cells in sub-G1 fraction. Nuclear fragmentation and AO/EtBr staining also consolidated that the major pathway of the programmed cell death was mediated through apoptosis. To understand the mechanism of apoptosis, the expression patterns of different key proteins involved in apoptosis were observed through immunoblot analysis.
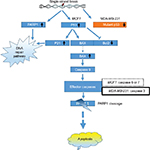
Increases in P53, as well as P21 protein, might lead to the cell cycle delay and apoptotic induction. Reports suggest that the reactive oxygen species (ROS) in SNP-treated cells are involved in anticancer activities.35–37 ROS-generated free radicals are known to induce DNA strand breaks.38 PARP1 plays an important role in repairing DNA single-strand breaks (SSBs), particularly in the absence of BRCA1. In the present study, upregulation of PARP1 protein suggests that GSNPs might induce SSBs in BRCA1 mutant MCF7 cells and MDA-MB-231 cells. PARP1 is activated at an intermediate stage of apoptosis and is inactivated by proteolytic cleavage at a late stage by caspase 3 and caspase 7. Such proteolytic cleavage is considered as a hallmark of apoptosis.39 We also observed the appearance of cleaved PARP1 bands, 24 h onward in MCF7 cells and 2 h onward in MDA-MB-231 cells, suggesting that GSNPs induced apoptosis in a time-dependent manner. MCF7 cells do not express caspase 3.40 Despite this, MCF7 cells undergo morphological apoptosis after treatment with a variety of agents and conditions.41,42 Liang et al,43 using a series of caspase inhibitors with overlapping specificities, enzyme-specific chromogenic substrates and an antibody specific for activated caspase 7, determined that apoptosis in MCF7 cells proceeds via sequential activation of caspases 9, 7 and 6. MDA-MB-231 cells have mutant P53 proteins due to the mutation at codon 280 of p53 gene (AGA to AAA), leading to the replacement of arginine to lysine.44 It is an established fact that the Bcl-2 family members, either inhibitor (Bcl-2) or activator (Bax), play an important regulatory role in apoptosis.45 In an earlier study, neocarzinostatin (NCS) treatment resulted in condensation and fragmentation of MCF7 cell nuclei and release of cytochrome C from the mitochondria to the cytosol. This apoptosis was accompanied by decreased levels of Bcl2 and increased levels of Bax.43 In another study, genistein-treated MDA-MB-231 cells exhibited downregulation of the mutant P53, whereas upregulation of P21 was observed. The authors of that study explained that the elevation of P21 protein is independent of P53 activation.44 In our present study, GSNPs also exhibited a similar expression pattern of Bcl2, Bax and cleaved caspase 9, indicating that Mentha arvensis-mediated GSNPs could also induce apoptosis in MCF7 and MDA-MB-231 cells by activating caspase 9. It is also clear that the effects of GSNPs are independent of the cellular ER and P53 statuses. The possible pathway of the mechanism of action is shown in Figure 17.
The Ames test is the golden assay to determine the mutagenic potential of medicinal plants/pharma products for an initial screening. A positive response in any single bacterial strain among four Salmonella mutants is sufficient to consider a substance as a mutagen.46 Any substance can be considered mutagenic if it produces at least a twofold concentration-dependent increase in the mean revertant colonies per plate in any one of these tester strains and there should be a dose–response relationship.47
Mentha arvensis-mediated GSNPs failed to elicit a mutagenic response in the Ames test in strains TA98, TA100, TA1535 and TA1538 of S. typhimurium. Considering the type of GSNPs, which are unlikely to be metabolized by S9, the experiment was conducted without the S9 metabolic activation system. The revertant colonies observed in four different doses clearly showed that the nanoparticles are nonmutagenic up to 5 mg per plate concentration (Table 1). The results were concluded based on the induced revertant colonies per plate with different concentrations. It was found that the revertants were not dose dependent, and none of the four concentrations showed double the number of revertants observed in the negative control. The mutagenicity test by the Ames test revealed that the Mentha arvensis-mediated GSNPs had no mutagenic potential toward all the four strains tested.
Conclusion
To the best of our knowledge, this is the first report that M. arvensis leaf extract can be effectively used to synthesize SNPs through a simple and efficient way. Synthesized GSNPs exhibited significant cytotoxicity to breast cancer (MCF7 and MDA-MB-231) cells at a comparable level with that of CSNPs. At equivalent doses, cytotoxicity to normal cells was significantly less. The size, shape and structure of GSNPs were confirmed by UV–Vis, XRD, FTIR, AFM and TEM analyses. Along with the efficient anticancer activity, GSNPs also demonstrated nonmutagenic attributes. Thus, GSNPs might be useful as a potential anticancer agent in breast cancer therapy.
Acknowledgments
The authors express their gratitude to Department of Biotechnology, India, for financial assistance through the grant no. BT/473/NE/TBP/2013, dated 13/02/2014. AB and SNH gratefully acknowledge the JRF and RA award of DBT, respectively. PPB is grateful to University Grants Commission (New Delhi) for the Meritorious Fellowship. SB is grateful to National Academy of Sciences, India for the Honorary Scientist award. The authors gratefully acknowledge the help of Professor Anupam Chatterjee, Department of Biotechnology and Bioinformatics, North-Eastern Hill University, Shillong, Meghalaya, for extending the FACS facility.
Disclosure
The authors report no conflicts of interest in this work.
References
Song JY, Kim BS. Rapid biological synthesis of silver nanoparticles using plant leaf extracts. Bioprocess Biosyst Eng. 2009;32(1):79–84. | ||
Jha AK, Prasad K, Prasad K, Kulkarni AR. Plant system: nature’s nanofactory. Colloids Surf B Biointerfaces. 2009;73(2):219–223. | ||
Konishi Y, Ohno K, Saitoh N, et al. Bioreductive deposition of platinum nanoparticles on the bacterium Shewanella algae. J Biotechnol. 2007;128(3):648–653. | ||
Willner I, Baron R, Willner B. Growing metal nanoparticles by enzymes. Adv Mater. 2006;18(9):1109–1120. | ||
Vigneshwaran N, Ashtaputre NM, Varadarajan PV, Nachane RP, Paraliker KM, Balasubramanya RH. Biological synthesis of silver nanoparticles using the fungus Aspergillus flavus. Mater Let. 2007;61(6):1413–1418. | ||
Mondal MK, Banerjee PP, Saha SK, et al. Selective reduction technique (SRT): a robust method to synthesize bioactive AG/Au doped grapheme oxide. Mater Des. 2016;102:186–195. | ||
Barua S, Banerjee PP, Sadhu A, et al. Silver nanoparticle as antibacterial and anticancer materials against human breast, cervical and oral cancer cells. J Nanosci Nanotechnol. 2016;16:1–9. | ||
Farooqui MA, Chauhan PS, Krishnamoorthy P, Shaik J. Extraction of silver nanoparticles from the leaf extracts of Clerodendrum inerme. Dig J Nanomater Biostruct. 2010;5(1):43–49. | ||
Puišo J, Jonkuvienė D, Mačionienė I, Šalomskienė J, Jasutienė I, Kondrotas R. Biosynthesis of silver nanoparticles using lingonberry and cranberry juices and their antimicrobial activity. Colloids Surf B Biointerfaces. 2014;121:214–221. | ||
Sadeghi B, Rostami A, Momeni SS. Facile green synthesis of silver nanoparticles using seed aqueous extract of Pistacia atlantica and its antibacterial activity. Spectrochim Acta A Mol Biomol Spectrosc. 2015;134:326–332. | ||
McGuire S. World cancer report 2014. Geneva, Switzerland: World Health Organization, international agency for research on cancer, WHO press, 2015. Adv Nutr. 2016;7(2):418–419. | ||
Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2014;136(5):E359–E386. | ||
Asthana S, Chauhan S, Labani S. Breast and cervical cancer risk in India: an update. Indian J Public Health. 2014;58(1):5–10. | ||
Sreedevi A, Javed R, Dinesh A. Epidemiology of cervical cancer with special focus on India. Int J Womens Health. 2015;7:405–414. | ||
Bhimba BV, Devi JS, Nandhini SU. Green synthesis and cytotoxicity of silver nanoparticles from extracts of the marine macroalgae Gracilaria corticata. Indian J Biotechnol. 2015;14:276–281. | ||
Vivek R, Thangam R, Muthuchelian K, Gunasekaran P, Kaveri K, Kannan S. Green biosynthesis of silver nanoparticles from Annona squamosa leaf extract and it’s in vitro cytotoxic effect on MCF-7 cells. Process Biochem. 2012;47(12):2405–2410. | ||
Durgawale P, Phatak R, Datkhile K, Hendre A, Durgawale P. Biologically synthesized silver nanoparticles from latex of Syandenium grantii and fresh leaves of Kalanchoe pinnata: potential source of cytotoxic agents against cervical cancer cells. Int J Sci Res. 2015;4(10):339–342. | ||
Kaba SI, Egorova EM. In vitro studies of the toxic effects of silver nanoparticles on HeLa and U937 cells. Nanotechnol Sci Appl. 2015;8:19–29. | ||
Miura N, Shinohara Y. Cytotoxic effect and apoptosis induction by silver nanoparticles in HeLa cells. Biochem Biophys Res Commun. 2009;390(3):733–737. | ||
AshaRani PV, Low Kah Mun G, Hande MP, Valiyaveettil S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano. 2009;3(2):279–290. | ||
Padmalochana K, Dhana Rajna MS. In-vitro anticancer activity of different extracts of Sesbania grandiflora against HEP2 cell lines. World J Pharm Sci. 2015;3(8):1589–1592. | ||
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. | ||
Bøyum A. Isolation of lymphocytes, granulocytes and macrophages. Scand J Immunol. 1976;5(s5):9–15. | ||
Tennant JR. Evaluation of the trypan blue technique for determination of cell viability. Transplantation. 1964;2(6):685–694. | ||
OECD. Guideline for the Testing of Chemical TG No. 471. Bacterial Reverse Mutation Test; 1997. Available from: http://www.oecd.org/dataoecd/18/31/1948418.pdf. Accessed March 10, 2017. | ||
Maron DM, Ames BN. Revised methods for the Salmonella mutagenicity test. Mutat Res. 1983;113(3–4):173–215. | ||
Kumar PPNV, Pammi SVN, Kollu P, Satyanarayana KVV, Shameem U. Green synthesis and characterization of silver nanoparticles using Boerhavia diffusa plant extract and their anti bacterial activity. Ind Crops Prod. 2014;52:562–566. | ||
Huang NM, Lim HN, Radiman S, et al. Sucrose ester micellar-mediated synthesis of Ag nanoparticles and the antibacterial properties. Colloids Surf A Biointerfaces. 2010;353(1):69–76. | ||
Basavaraja S, Balaji SD, Lagashetty A, Rajasab AH, Venkataraman A. Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium semitectum. Mater Res Bull. 2008;43(5):1164–1170. | ||
Begum NA, Mondal S, Basu S, Laskar RA, Mandal D. Biogenic synthesis of Au and Ag nanoparticles using aqueous solutions of Black Tea leaf extracts. Colloids Surf B Biointerfaces. 2009;71(1):113–118. | ||
Narayanan KB, Sakthivel N. Extracellular synthesis of silver nanoparticles using the leaf extract of Coleus amboinicus Lour. Mater Res Bull. 2011;46(10):1708–1713. | ||
Sangiliyandi G, Jagadeesh R, Sri Nurestri AM, Priscilla AJ, Sabaratnam V. Green synthesis of silver nanoparticles using Ganoderma neojaponicum Imazeki: a potential cytotoxic agent against breast cancer cells. Int J Nanomedicine. 2013;8:4399–4413. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Das J, Das MP, Velusamy P. Sesbania grandiflora leaf extract mediated green synthesis of antibacterial silver nanoparticles against selected human pathogens. Spectrochim Acta A Mol Biomol Spectrosc. 2013;104:265–270. | ||
Wan J, Liu T, Mei L, et al. Synergistic antitumour activity of sorafenib in combination with tetrandrine is mediated by reactive oxygen species (ROS)/Akt signaling. Br J Cancer. 2013;109(2):342–350. | ||
Whibley CE, McPhail KL, Keyzers RA, et al. Reactive oxygen species mediated apoptosis of esophageal cancer cells induced by marine triprenyl toluquinones and toluhydroquinones. Mol Cancer Ther. 2007;6(9):2535–2543. | ||
Ravid A, Koren R. The role of reactive oxygen species in the anticancer activity of vitamin D. Recent Results Cancer Res. 2003;164:357–367. | ||
Podder S, Chattopadhyay A, Bhattacharya S, Ray MR, Chakraborty A. Fluoride induced genotoxicity in mouse bone marrow cells: effect of buthionine sulfoximine and N-acetyl-L-cysteine. J Appl Toxicol. 2010;31(7):618–625. | ||
Soldani C, Lazzè MC, Bottone MG, et al. Poly (ADP-ribose) polymerase cleavage during apoptosis: when and where? Exp Cell Res. 2001;269(2):193–201. | ||
Jänicke RU, Sprengart ML, Wati MR, Porter AG. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J Biol Chem. 1998;273(16):9357–9360. | ||
Eck-Enriquez K, Kiefer TL, Spriggs LL, Hill SM. Pathways through which a regimen of melatonin and retinoic acid induces apoptosis in MCF-7 human breast cancer cells. Breast Cancer Res Treat. 2000;61(3):229–239. | ||
Oberhammer F, Wilson JW, Dive C, et al. Apoptotic death in epithelial cells: cleavage of DNA to 300 and/or 50 kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 1993;12(9):3679–3684. | ||
Liang Y, Yan C, Schor NF. Apoptosis in the absence of caspase 3. Oncogene. 2001;20(45):6570–6578. | ||
Yiwei LI, Upadhyay S, Bhuiyan M, Sarkar FH. Induction of apoptosis in breast cancer cells MDA MB 231 by genistein. Oncogene. 1999;18:3166–3172. | ||
Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9(1):47–59. | ||
Zeiger E. Mutagens that are not carcinogens: faulty theory or faulty tests? Mutat Res. 2001;492(1):29–38. | ||
Katzer A, Hockertz S, Buchhorn GH, Loehr JF. In vitro toxicity and mutagenicity of CoCrMo and Ti6Al wear particles. Toxicology. 2003;190(3):145–154. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.