Back to Journals » OncoTargets and Therapy » Volume 13
MBD2 Correlates with a Poor Prognosis and Tumor Progression in Renal Cell Carcinoma
Authors Li L , Li N, Liu N , Huo F, Zheng J
Received 31 March 2020
Accepted for publication 27 August 2020
Published 7 October 2020 Volume 2020:13 Pages 10001—10012
DOI https://doi.org/10.2147/OTT.S256226
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjay Singh
Liantao Li,1– 4,* Na Li,2,3,* Nianli Liu,1,4 Fuchun Huo,5 Junnian Zheng1,2
1Cancer Institute, Xuzhou Medical University, Xuzhou 221000, People’s Republic of China; 2Center of Clinical Oncology, Affiliated Hospital of Xuzhou Medical University, Xuzhou 221000, People’s Republic of China; 3Department of Radiation Oncology, Affiliated Hospital of Xuzhou Medical University, Xuzhou 221000, People’s Republic of China; 4Jiangsu Center for the Collaboration and Innovation of Cancer Biotherapy, Xuzhou 221000, People’s Republic of China; 5Department of Pathology, Xuzhou Medical University, Xuzhou 221000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Junnian Zheng 84 West Huaihai Road, Xuzhou, Jiangsu, People’s Republic of China
Tel +8605168558230
Email [email protected]
Purpose: DNA methylation plays an important role in regulating gene expression. Methyl-CpG-binding domain (MBD) proteins recognize and bind to methylated DNA, which mediate gene silencing by the interaction with deacetylases and histone methyltransferases. MBD2 has been reported in various human cancers; however, its clinical implication and potential regulatory role in renal cell carcinoma (RCC) have not been elaborated.
Materials and Methods: In the study, we estimated the expression and prognostic value of MBD2 in RCC cell lines and tissues by Western blotting and immunohistochemistry. The associations of MBD2 expression and pathological characters and survival in RCC patients were performed using χ 2 and Kaplan–Meier survival analysis, respectively. Univariate and multivariable Cox regression analyses suggested the independent predictors in RCC prognosis. The functional role of MBD2 in RCC progression was assessed by in vitro cell experiments. In addition, we identified the MBD2-mediated alterations of protein-related proliferation and EMT markers in RCC cells after MBD2 overexpression and knockdown.
Results: We found that the protein levels of MBD2 were upregulated in RCC cells and tissues. High MBD2 expression was related to TNM stage and predicted poorer survival in RCC. Enforced expression of MBD2 significantly promoted the proliferation, cycle progress, invasion and migration of RCC cells in vitro. However, downregulating MBD2 remarkably weakened the above cell functions. Mechanistically, the promotive effect of MBD2 overexpression may be regulated by its effects onp21, p53 and Cyclin D1 expression and EMT process.
Conclusion: These results indicated that MBD2confers an oncogenic function in the malignant progression of RCC. MBD2 could be served as a meaningful prognostic biomarker and a latent therapeutic target in RCC patients.
Keywords: MBD2, RCC, prognosis
Introduction
RCC is one of the most common malignant tumors in the urinary system.1 It is one of the few tumors with a higher incidence in males than in females and tends to increase worldwide.2 Due to the concealed onset of RCC and the absence of typical clinical symptoms in the early stage, about 30% of patients have metastasis and malignant progression at the time of RCC diagnosis,3,4 and postoperative recurrence occurs in 20%-30% of patients.5 Typical RCC triad (low back pain, hematuria, abdominal mass) occurs or does not occur in advanced patients, among those who are insensitive to radiotherapy and conventional chemotherapy and have no significant clinical effects on treatments. Therefore, further understanding of the tumorigenesis of RCC is an unmet clinical need for identifying reliable prognostic biomarkers and therapeutic targets for RCC patients.
DNA methylation regulates the transcriptional activity of DNA without permanent changes of nucleotide sequences. Epigenetic information of DNA methylation is interpreted and translated by recruiting “reader” molecules known as methyl-binding proteins (MBPs).6–8 MBPs binds to methylated DNA, namely CpG dinucleotides, and can organically link the DNA methylation with histone modification.6–9 Methyl-CpG-binding domain (MBD)-containing proteins belong to the first family of MBPs. Among these members, MBD2 evinces the deepest affinity with methylated DNA, which mimics chromatin remodeling or modification factors, alters the structure of chromatin and interferes with the binding of transcription factors to target genes following its binding to methylated DNA, subsequently inhibiting gene transcription concomitant with the alterations of histone conformation.6,10–12MBD2 is identified as a multiexon gene and serves as a transcriptional repressor/activator depending on the cellular context.13 MBD2 protein is composed of 411 amino acids, among which ranging from 145 to 213 amino acids is the methyl-binding domain, which can bind to the specific methylation sequences of target genes.
Aberrant DNA methylation modifications are frequently detected in various tumors, and the main mechanisms for DNA methylation-involved tumorigenesis are that methylation levels of the promoter region of anti-oncogene are elevated, which promotes the key anti-oncogene silencing and drives tumorigenesis.12,14–16 MBD2 has been reported in multiple human malignancies, including gastric cancer,17 breast cancer,18,19 colorectal cancer,20 glioblastoma,21,22 hilar cholangiocarcinoma,23 hepatocellular carcinoma,24,25 chronic myeloid leukemia26 and prostate cancer.27 Previous studies confirmed that MBD2 mediates the transcriptional repression of tumor suppressor genes, such as hTERT,28 GSTP1,29 BAI1,21 p14ARF/p16INK4a,30 and 14-3-3sigma,27 which supports the pivotal role of MBD2 in abnormal epigenetic regulation of tumors. MBD2 is recruited to the promoter of BAI1 considered as an important tumor suppressor of antiangiogenic activity and maintains its epigenetic silencing to promote the carcinogenesis and tumor progression of glioblastoma.21 miR-520b impedes glucose metabolism, invasion, angiogenesis and chemosensitivity of glioma cells by directly targeting MBD2.22 MBD2 deletion induces the activation of JAK2/STAT3 signaling pathway and inhibits tumor growth in chronic myeloid leukemia.26 Increased MBD2 is proposed as an independent prognostic factor for overall (OS) and disease-free survival (DFS) for hepatocellular carcinoma patients.24 LOC105369748 upregulates MBD2 through binding to miR-5095, which conveys oncogenic signals to accelerate the carcinogenesis and progression of hepatocellular carcinoma.25 All above evidence indicates an oncogenic role of MBD2 in the initiation and progression of cancers. However, the pathophysiologic function and regulatory mechanism of MBD2 in RCC remain largely unknown.
In the present report, we sought to ascertain the clinical and prognostic significance of MBD2 in RCC tissues and discovered that highly expressed MBD2 was associated with the pathological characteristics and prognosis of RCC patients. Furthermore, effects of MBD2 on proliferation, cell cycle, migration and invasion were investigated in RCC cells, which provided evidence that enforced expression of MBD2 promoted cell proliferation, cycle progression, migration and invasion in vitro. Mechanistically, MBD2 regulated proteins related to cell proliferation and EMT markers. Taken together, we reported that MBD2 may be an independent prognostic indicator of RCC, which suggested the therapeutic potential of targeting MBD2 for RCC patients.
Materials and Methods
Patients and Tissue Samples
A total of 109 pairs of RCC tissue samples and matched normal renal tissues were collected from the patients who received surgical resection of primary RCC at the Affiliated Hospital of Xuzhou Medical University (Xuzhou, China) from December 2013 to November 2015. All patients selected in the study were histopathological diagnoses with RCC and had never been subjected to radiotherapy and/or chemotherapy preoperatively. The detailed clinical and pathologic information of RCC patients were recorded and stored completely. The clinicopathological parameters included age, gender, Fuhrman grade, TNM stage. All RCC patients were termly followed up from 1 to 65 months to assess postoperative survival, and follow-up information was obtained by reviewing the well-documented medical records. After collection, all fresh tissues were quickly stored in liquid nitrogen and frozen at −80°C for subsequent experiments. The study protocol involving human specimens was approved by the Review Board of the Affiliated Hospital of Xuzhou Medical University. Written informed consent was collected from all participants prior to the study.
Immunohistochemistry Staining and Evaluation
Immunohistochemical (IHC) analysis was performed according to our previously described methods.31 Briefly, formalin-fixed, paraffin-embedded RCC tissues were cut into 4 μm sections. After 2 h baking, the section was deparaffinized with dimethylbenzene and rehydrated in graded ethanol solutions, respectively. Then, the section was heated at 95°C for 5 min in a citrate buffer (pH = 6.0) for antigen retrieval. For blocking endogenous peroxidase activity, the slide was washed with PBS twice and then treated with 3% hydrogen peroxide for 15 min. After the incubation of normal goat serum, the section was incubated with the anti-MBD2 (Abcam, Cambridge, MA, US) for 12 h at 4°C. Then, the slide was incubated with appropriate secondary antibody for 1 h at room temperature. The detection of antigenic staining was conducted by 3, 3′-diaminobenzidine (DAB; Beyotime Biotechnology, Shanghai, China) and counterstained with hematoxylin (Beyotime Biotechnology). Finally, the slide was dehydrated and sealed.
Two independent pathologists assessed the IHC staining in each section by light microscopic examination. Staining intensity was assigned as follows: 0 (-); 1 (+); 2 (++); 3 (+++) and positively stained area was assigned as follows: 1 (0–25%); 2 (26–50%); 3 (51–75%); 4 (76–100%). Staining pattern of each section was scored according to the immunoreactive score (IRS) determined by the product of staining intensity and positively stained area. All specimens were categorized into low MBD2 expression (IRS < 6) and high MBD2 expression (IRS ≥ 6).
Cell Culture
Human RCC cell lines including 786-O, ACHN, 769-P, Caki-1 and Caki-2 and normal human renal tubular epithelial cell HK-2 were supplied by the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured according to our previously described methods.31
Transfection
Small interference RNAs (siRNAs) targeting human MBD2 (siMBD2) and negative controls (siCon) were from GenePharma (Shanghai, China). The siRNA sense sequences were 5ʹ-UUACUAGGCAUCAUCUUUCUU-3ʹ for siMBD2 and 5ʹ-GCCUUGGCAGCCUAGGCGA-3ʹ for siCon.32 siRNAs transfection in RCC cells was performed using siLentFect™ Lipid Reagent (Bio-Rad, Hercules, CA, USA). For the overexpression ofMBD2, human MBD2 plasmid was subcloned into the pcDNA3.1 vector (GenePharma). pcDNA3.1-MBD2 plasmids and empty vectors were transfected into the RCC cells using Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA). Cells were cultured for 24 h or 48 h after transfection and then used for the following experiments.
Western Blotting
Cells were lysed in the cold RIPA buffer (Beyotime Biotechnology) with Phenylmethylsulfonyl fluoride (PMSF), and protein contents were determined by the BCA assay (BCA; Beyotime Biotechnology). Equal amount of protein was separated by 10% SDS-PAGE gel and then electrophoretically transferred to nitrocellulose membranes. The membrane was blocked with 5% non-fat skim milk for 1 h and then probed overnight at 4°C with primary antibodies: MBD2 (Abcam), E-cadherin (Abcam), p21 (Abcam), p53 (Abcam), N-cadherin (Abcam), Vimentin (Abcam), Cyclin D1 (Abcam) and GAPDH (Zhongshan biotech, Beijing, China). The following day, membrane was incubated with secondary antibodies. Finally, the signal of protein was visualized by an enhanced chemiluminescence detection system.
Cell Proliferation Assay
Cell proliferation was measured by the Cell Counting Kit-8 reagent (Dojindo, Kumamoto, Japan). Transfected 786-O and ACHN cells were seeded into 96-well plates at a density of 2 × 103 cells and incubated at 37°C. After the inoculation for indicated times, 10 μL CCK-8 reagent was added into wells containing 100 μL of serum-free medium. The absorbance at 450 nm (OD 450) of cells was measured by a microplate reader.
Transwell Assay
Transwell assay was used to assess the cell invasion and migration ability using transwell chamber (BD Bioscience, USA). For invasion assay, matrigel was pre-coated in the upper chamber. 1x105 transfected cells in 100 μL of medium without FBS were seeded into the upper chamber, while the medium with 10% FBS (600 μL) was added into the bottom of the chambers. After 24 h, cells on the membrane filters were fixed in cold methanol, and noninvasive cells were softly erased with a cotton swab. Finally, cells adhered to the lower membrane were stained with the crystal violet, calculated and photographed using a bright field microscopy at 200x magnification.
Cell Cycle Assay
Transfected RCC cells were harvested, washed, then fixed in 70% ice-cold ethanol for 12 h. Cells were then resuspended with DNA-staining solution for 30 min at room temperature in the dark. Finally, samples were detected using a FACScan flow cytometer (BD Biosciences). The distribution of cell cycle was determined using the CellQuest software program (BD Biosciences).
Statistical Analysis
All statistical analyses were determined using the SPSS 22.0. Data were presented as means ± SD. The statistical significance of different groups was compared by t-test or one-way ANOVA. The Χ2 test was employed to assess the association between MBD2 expression and clinicopathological features of RCC, and the Kaplan–Meier method and Log rank test were used to estimate the OS and DFS. The significance of MBD2 expression in RCC patients was examined by the univariate and multivariate Cox regression analyses. P-value <0.05 was considered statistically significant.
Results
MBD2 is Highly Expressed in RCC
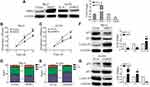
Initially, MBD2 expression was detected in RCC cell lines and tissues using Western blotting. Results showed that MBD2 significantly elevated in the RCC cell lines including 786-O, ACHN, 769-P, Caki-1 and Caki-2 compared with normal human renal tubular epithelial cell HK-2 (Figure 1A, P < 0.001). Next, we analyzed MBD2 expression through excavating the online server GEPIA based on TCGA and GTEx data (http://gepia.cancer-pku.cn/detail.php?gene=MBD2) and found that MBD2 was frequently overexpressed in the RCC tissues compared with adjacent non-tumor tissues (Figure 1B). We also detected the relative expression of MBD2 between RCC tissues and tumor-adjacent tissues by available frozen specimens randomly selected from 109 cases of RCC. Comparative analysis revealed that the protein levels of MBD2 were higher in RCC tissues than in matched normal tissues (Figure 1C, P < 0.05). To further investigate the MBD2 expression in RCC tissues, IHC staining was performed with 109 tissue specimens. Strong staining of MBD2 protein was frequently observed in RCC tissues, while weak or negative staining was more prone to be shown in adjacent non-tumor tissues. Representative illustrations for MBD2 staining of both RCC and normal tissues are shown in Figure 1D and Supplementary Figure 1A. According to the quantitative analysis of MBD2 staining, RCC specimens were divided into two categories: low MBD2 expression (IRS: 0–4) and high MBD2 expression (IRS: 6–12). Sixty-six of 109 (60.6%) RCC samples were exhibited high MBD2 expression; however, the high expression in tumor-adjacent tissues was merely 17 of 109 (15.6%). In addition, MBD2 staining was detected in both cytoplasm and nucleus. Taken together, these results suggested MBD2 was elevated in RCC and may be associated with RCC progression.
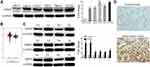 |
Figure 1 MBD2 expression is upregulated in RCC cells and tissues. (A) The protein expression of MBD2 was determined in the several human RCC cell lines (Caki-1, Caki-2, 769-P, 786-O and ACHN) and human normal tubular epithelial cell line (HK-2) by Western blotting. (B) The expression levels of MBD2 in KIRC (Kidney renal clear cell carcinoma; (T) and adjacent normal (N) tissues from TCGA and GTEx data were analyzed by excavating the online server GEPIA (http://gepia.cancer-pku.cn/detail.php?gene=MBD2). (C) MBD2 protein levels in 6 representative pairs of RCC (T) and adjacent normal (N) tissues were presented in the immunoblot. (D) The representative IHC staining of MBD2 in RCC and normal renal tissues. Original magnification, × 400. Data represents the means ± SD. *P < 0.05; **P < 0.01; ***P < 0.001. |
MBD2 is Correlated with Poor Prognosis in RCC
The relationship between MBD2 expression and clinicopathological findings was presented (Table 1). Elevated MBD2expression was significantly correlated with T stage (P = 0.044), distant metastasis (P = 0.016) and TNM stage (P = 0.011), whereas no compelling correlation was found in age, gender, Fuhrman grade and lymph node involvement.
 |
Table 1 The Correlations Between Expression and Clinicopathologic Characteristics in RCC Patients |
For further understanding the effect of MBD2 expression on the clinical outcomes of RCC patients, we initially effectuated the survival curves by online server GEPIA, which revealed that high MBD2 expression predicted the better OS (Figure 2A, P < 0.001) and DFS (Figure 2B, P < 0.05). Nevertheless, we performed the Kaplan–Meier survival curves and Log rank test from the 109 RCC samples, and results indicated that the RCC patients with high MBD2 expression had poorer 5-year OS and DFS after radical nephrectomy than these with low MBD2 expression (Figure 2C for OS, P < 0.001; Figure 2D for DFS, P < 0.001). We also conducted univariate and multivariate analyses through a Cox proportional hazard model to explore the prognostic factors in RCC patients. Univariate analysis uncovered that T stage, lymph node involvement, TNM stage and MBD2 expression were identified as prognostic factors for OS and DFS (Tables 2 and 3), while age, Fuhrman grade and distant metastasis were not associated with the OS and DFS. Moreover, further multivariate analysis confirmed that increased MBD2 expression was an independent risk factor for the OS and DFS in RCC patients (Table 2, HR: 3.004, 95% CI: 1.519–4.149, P < 0.001 for OS; Table 3, HR: 2.511, 95% CI: 1.519–4.149, P < 0.001 for DFS). Collectively, our results indicated that MBD2 is involved in RCC progression and may be proposed as a valuable prognostic marker for RCC patients.
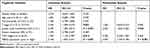 |
Table 2 Univariate and Multivariate Cox Analyses of Prognostic Markers for OS in RCC Patients |
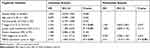 |
Table 3 Univariate and Multivariate Cox Analysis of Prognostic Markers for DFS in RCC Patients |
The Effects of MBD2 on the Proliferation of RCC Cells
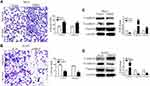
Given that increased MBD2 expression was related to malignant progression and poorer prognostic, we hypothesized that MBD2 function as an oncogene in RCC cells. To decipher this, we performed the gain-of-function and loss-of-function experiments. Initially, 768-O and ACHN cells were transfected with MBD2 plasmid or siRNA MBD2, and the transfection efficiency of MBD2 overexpression and knockdown are shown Figure 3A (P < 0.01). CCK-8 assays were utilized to determine the RCC cell activity in vitro. Enforced MBD2 expression accelerated the cellular growth in 768-O cells (Figure 3B, P < 0.01). Inversely, the blockade of MBD2 expression restrained the proliferative ability of ACHN cells (Figure 3C, P < 0.01). Meanwhile, cell cycle assay was applied to analyze the MBD2-mediated cell cycle profiles. MBD2 amplification compelled the cycle transition from G1 phase to S phase in 768-O cells (Figure 3D, Supplementary Figure 1B). MBD2 silencing significantly decelerated cycle transition and increased G1 phase populations in ACHN cells (Figure 3E, Supplementary Figure 1B).
Malignant proliferation is an important characteristic of tumor cells, which is concomitant with the disorders of cell cycle-related proteins and signaling pathways. Previous studies have reported the promotive proliferation role of MBD2 glioma cells.22 Western blotting revealed that MBD2 potentiated the Cyclin D1 expression concomitant with the inhibition of p53 and p21 in 768-O cells (Figure 3F, P < 0.01). Inverse results were observed in the ACHN cells with MBD2 silencing (Figure 3G, P < 0.05). Overall, MBD2 promoted the expression of cyclin D1 and suppressed the expression of p53 and p21, leading to the proliferation of RCC cells.
The Effects of MBD2 on Migration and Invasion of RCC Cells
IHC results confirmed the potential oncogene role of MBD2 and its overexpression was associated with aggressive phenotypes in RCC. We next investigated the MBD2-mediated the migration and invasion of RCC cells through quantification analysis of transwell assay. Results indicated that the 768-O cells transfected with MBD2 plasmid obviously increased the cell migratory and invasive capabilities (Figure 4A, P < 0.01). The MBD2 silencing conferred by siRNA effectively impaired the number of invaded ACHN cells (Figure 4B, P < 0.001).
Tumor cells are characterized by the local infiltration and distant metastasis. EMT is a process in which polar epithelial cells transform into motile mesenchymal cells and acquire the ability to migration and invasion. We explored the alterations of EMT markers in RCC cells after MBD2 treatment. Western blotting showed that the elevated expression of MBD2 was accompanied by an increase in mesenchymal markers, N-cadherin and Vimentin, whereas a decline in epithelial marker, E-cadherin (Figure 4C, P < 0.01). Abating expression of MBD2 totally produced an opposite effect (Figure 4D, P < 0.01). Together, MBD2 potentiated the migration and invasion of RCC cells and induced EMT.
Discussion
DAN methylation is widely existed in bacteria, plants and mammal, which is involved in maintaining the stability of genome and chromosome structure, gene imprinting, X chromosome inactivation and gene silencing and is closely related to the occurrence and development of cell carcinogenesis and cancer.6,9,13,33–35 DNA methylation can directly or indirectly affect the gene transcription and expression without the alterations of DNA sequences and gene products. Researches show that abnormal DNA methylation is involved in tumorigenesis by affecting the structure of chromatin and the expression of tumor suppressor genes and oncogenes.33,36 The abnormal methylation of tumor-related genes exists in almost all human tumors. In cancers, the hypermethylation of CpG islands in the promoter region of tumor suppressor genes, as a universal inactivation mechanism, mediates the silencing of tumor suppressor genes, which is involved in the carcinogenesis and tumor progression.10,16,21,35
MBD family plays an important role in the processes of aberrant DNA methylation related to tumors. As a member of MBD family, the MBD2, a methylation-dependent transcriptional repressor, is characterized by specifically recognizing and binding to methylated DNA and inhibits gene transcription through recruiting co-repressors like the NuRD complex to methylated CpG islands.12,37 Abundant evidence has demonstrated that MBD2 mediates the transcriptional repression of tumor suppressor genes, such as hTERT,28 GSTP1,29 BAI1,21 p14ARF/p16INK4a,30 14-3-3sigma.27 Amplified MBD2 is detected in glioblastoma21,22 and hepatocellular carcinoma.38 The MBD2-mediated epigenetic silencing of BAI1 drives tumor growth in glioblastoma.21 MBD2 deletion induces the inactivation of JAK2/STAT3 pathway and suppresses cell proliferation capacity in chronic myeloid leukemia.26 MBD2−/- is protective against the carcinogenesis and tumor progression and significantly suppresses the intestinal tumorigenesis in ApcMin/+ mice.39 Devailly et al reported that MBD2 is enriched in the methylated regions of target genes and associated with the transcriptional silencing during malignant transformation of human mammary cells.18 However, the expression pattern and functional roles of MBD2 in RCC pathogenesis remain yet reported.
In order to illuminate the biological and clinical significance of MBD2, we firstly detected the protein levels of MBD2 in RCC cells and tissues. Western blotting showed that increased MBD2 was found in the RCC cell lines compared with normal renal epithelial cell HK-2. And high expression profiles of MBD2 were shown in the RCC tissues when compared with adjacent normal tissues. In addition, we performed the IHC staining with 109 RCC specimens, which supported the increased MBD2 in RCC. Higher expression of MBD2 was significantly associated with T stage, distant metastasis, and TNM stage. Survival analysis revealed that MBD2 expression was negatively correlated with the OS and DFS of RCC patients, and both univariate and multivariate analysis demonstrated the role of MBD2 as an independent prognosis predictor for RCC patients. In accordance with this, Liu et al reported that the upregulated MBD2expression, as a candidate prognostic factor in hepatocellular carcinoma, was associated with poor OS and DFS, along with the clinicopathological characteristics including tumor size, vascular invasion and BCLC stage. Increased MBD2 is associated with malignant progression of glioma tissues.21,22 In contrast, several studies have yielded the conflicting results as to the MBD2 expression in cancer and matched normal tissues.17,40
We studied the biological effects of MBD2 gene in RCC cells via interfering and overexpressing MBD2. Our results suggested that MBD2 contributed to RCC cells malignancy. Forced expression of MBD2 apparently potentiated the proliferation, migration and invasion of RCC cells through CCK-8 and transwell assays. Meanwhile, MBD2 expedited the cell cycle progression from G1 phase to S phase. On the contrary,MBD2 depletion impeded cell proliferation, migration and invasion and decelerated cell cycle progression. Western blotting indicated that the aberrant expression of MBD2 augmented the Cyclin D1 level and impaired the p53 and p21 levels in RCC cells. CyclinD1 activates and binds to the cell cycle-dependent kinase 4/6 (CDK4/6) to form a kinase complex withCyclin D1 as a regulatory subunit and CDK4/6 as catalytic subunit, which enables cells to enter the S phase through G1/S regulatory point.41 Tumor suppressor gene p21 is a broad-spectrum inhibitor of cell CDK and a negative regulator of cell cycle, which can prevent the cells from passing through the G1-S phase. p53 bind to the promoter region of p21 to activate its transcription, preventing the transition from G1 to S phase and arresting the cell to G1 phase,42 Therefore, MBD2 may regulate the proliferation ability of RCC cells partly through Cyclin D1, p53and p21. Moreover, MBD2 increased the mesenchymal markers expression and decreased the epithelial marker (E-cadherin) expression in RCC cells. EMT characterized by the deletion of epithelial cell phenotypes and the appearance of mesenchymal cell phenotypes works as a decisive role in the tumorigenesis, invasion and metastasis.43,44 EMT enhances the migration and invasion of cancer cells by deleting the expression of connective molecules, potentiating the adhesion and motor ability.43,44 MBD2 may regulate the migration and invasion partly via EMT molecules, E-cadherin, N-cadherin and Vimentin. Together, MBD2 conferred on cells tumor malignant properties, promoted the cell proliferation, cycle progress, migration and invasion and regulates the proliferation and metastasis-related proteins in RCC, which is consistent with the previous reports of supporting cancer-promoting role of MBD2 in glioma,22 hepatocellular carcinoma,24 chronic myeloid leukemia.26 The exact molecular mechanisms of MBD2 affecting the proliferation, migration and invasion of RCC cells need to be further explored.
Conclusion
In conclusion, the present study provides evidence for the connection between MBD2 and RCC. We conclude that MBD2 shows a high expression in both RCC tissues and cell lines, is associated with a poor prognosis of RCC patients and might be an independent prognostic factor for RCC patients. MBD2 acts as an oncogene in RCC cells by promoting the cell growth, cycle progress, migration and invasion. Further researches are still in demand to clarify the precise molecular mechanism of MBD2 in RCC. Therefore, targeting MBD2 would be considered as a potential therapeutic avenue for RCC patients.
Author Contributions
Junnian Zheng was involved in the study design. Liantao Li was a major contributor in writing the manuscript. Li Na, Liantao Li and Nianli Liu analyzed the patient data. Li Na and Fuchun Huo conducted the in vitro experiments. All authors approved the final manuscript. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Funding
The present study was financially supported by the Jiangsu Provincial Medical Youth Talent (QNRC2016773), Jiangsu Provincial Science and Technology Program (BK20161157), Six Talent Peaks Project in Jiangsu Province (WSN-119), Post graduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_2185), the Jiangsu Provincial Medical Innovation Team under Grant CXTDA2017034.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387(10021):894–906. doi:10.1016/S0140-6736(15)00046-X
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30.
3. Capitanio U, Bensalah K, Bex A, et al. Epidemiology of renal cell carcinoma. Eur Urol. 2019;75(1):74–84. doi:10.1016/j.eururo.2018.08.036
4. Ljungberg B, Bensalah K, Canfield S, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol. 2015;67(5):913–924. doi:10.1016/j.eururo.2015.01.005
5. Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009.
6. Berger J, Bird A. Role of MBD2 in gene regulation and tumorigenesis. Biochem Soc Trans. 2005;33(Pt 6):1537–1540. doi:10.1042/BST0331537
7. Jablonska E, Reszka E. Selenium and epigenetics in cancer: focus on DNA methylation. Adv Cancer Res. 2017;136:193–234.
8. Jeltsch A, Broche J, Lungu C, Bashtrykov P. Biotechnological Applications of MBD Domain Proteins for DNA Methylation Analysis. J Mol Biol. 2020;432:1816–1823.
9. Sarraf SA, Stancheva I. Retraction notice to: methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2019;73(5):1084. doi:10.1016/j.molcel.2019.02.023
10. Ginder GD, Williams DC. Corrigendum to “Readers of DNA methylation, the MBD family as potential therapeutic targets” [Pharmacology & Therapeutics 184 (1) (2018) 98–111]. Pharmacol Ther. 2018;190:237–238. doi:10.1016/j.pharmthera.2018.09.006
11. Detich N, Theberge J, Szyf M. Promoter-specific activation and demethylation by MBD2/demethylase. J Biol Chem. 2002;277(39):35791–35794. doi:10.1074/jbc.C200408200
12. Parry L, Clarke AR. The roles of the methyl-CpG binding proteins in cancer. Genes Cancer. 2011;2(6):618–630. doi:10.1177/1947601911418499
13. Mahmood N, Rabbani SA. DNA methylation readers and cancer: mechanistic and therapeutic applications. Front Oncol. 2019;9:489.
14. Zhao X, Yang F, Li S, et al. CpG island methylator phenotype of myelodysplastic syndrome identified through genome-wide profiling of DNA methylation and gene expression. Br J Haematol. 2014;165(5):649–658. doi:10.1111/bjh.12811
15. Liu Y, Liu Y, Pan X, Li SC, Yang S, Li SC. DNA methylation markers for pan-cancer prediction by deep learning. Genes. 2019;10(10):778. doi:10.3390/genes10100778
16. Mahmood N, Rabbani SA. Targeting DNA hypomethylation in malignancy by epigenetic therapies. Adv Exp Med Biol. 2019;1164:179–196.
17. Pontes TB, Chen ES, Gigek CO, et al. Reduced mRNA expression levels of MBD2 and MBD3 in gastric carcinogenesis. Tumour Biol. 2014;35(4):3447–3453. doi:10.1007/s13277-013-1455-y
18. Devailly G, Grandin M, Perriaud L, et al. Dynamics of MBD2 deposition across methylated DNA regions during malignant transformation of human mammary epithelial cells. Nucleic Acids Res. 2015;43(12):5838–5854. doi:10.1093/nar/gkv508
19. Teslow EA, Mitrea C, Bao B, et al. Obesity-induced MBD2_v2 expression promotes tumor-initiating triple-negative breast cancer stem cells. Mol Oncol. 2019;13(4):894–908. doi:10.1002/1878-0261.12444
20. Yuan K, Xie K, Fox J, et al. Decreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in mice. Gastroenterology. 2013;145(4):853–864. doi:10.1053/j.gastro.2013.06.008
21. Zhu D, Hunter SB, Vertino PM, Van Meir EG. Overexpression of MBD2 in glioblastoma maintains epigenetic silencing and inhibits the antiangiogenic function of the tumor suppressor gene BAI1. Cancer Res. 2011;71(17):5859–5870. doi:10.1158/0008-5472.CAN-11-1157
22. Cui S, Liu L, Wan T, Jiang L, Shi Y, Luo L. MiR-520b inhibits the development of glioma by directly targeting MBD2. Am J Cancer Res. 2017;7(7):1528–1539.
23. Chen Y, Luo J, Tian R, Sun H, Zou S. miR-373 negatively regulates methyl-CpG-binding domain protein 2 (MBD2) in hilar cholangiocarcinoma. Dig Dis Sci. 2011;56(6):1693–1701. doi:10.1007/s10620-010-1481-1
24. Liu W, Wang N, Lu M, Du XJ, Xing BC. MBD2 as a novel marker associated with poor survival of patients with hepatocellular carcinoma after hepatic resection. Mol Med Rep. 2016;14(2):1617–1623. doi:10.3892/mmr.2016.5404
25. Li C, Li H, Yang J. Silencing of a novel lncRNA LOC105369748 suppresses the progression of hepatocellular carcinoma by sponging miR-5095 from MBD2. J Cell Physiol. 2019;234(10):18504–18512. doi:10.1002/jcp.28486
26. Cheng L, Tang Y, Chen X, et al. Deletion of MBD2 inhibits proliferation of chronic myeloid leukaemia blast phase cells. Cancer Biol Ther. 2018;19(8):676–686. doi:10.1080/15384047.2018.1450113
27. Pulukuri SM, Rao JS. CpG island promoter methylation and silencing of 14-3-3σ gene expression in LNCaP and Tramp-C1 prostate cancer cell lines is associated with methyl-CpG-binding protein MBD2. Oncogene. 2006;25(33):4559–4572. doi:10.1038/sj.onc.1209462
28. Chatagnon A, Bougel S, Perriaud L, Lachuer J, Benhattar J, Dante R. Specific association between the methyl-CpG-binding domain protein 2 and the hypermethylated region of the human telomerase reverse transcriptase promoter in cancer cells. Carcinogenesis. 2009;30(1):28–34. doi:10.1093/carcin/bgn240
29. Bakker J, Lin X, Nelson WG. Methyl-CpG binding domain protein 2 represses transcription from hypermethylated pi-class glutathione S-transferase gene promoters in hepatocellular carcinoma cells. J Biol Chem. 2002;277(25):22573–22580. doi:10.1074/jbc.M203009200
30. Magdinier F, Wolffe AP. Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc Natl Acad Sci U S A. 2001;98(9):4990–4995. doi:10.1073/pnas.101617298
31. Huo FC, Pan YJ, Li TT, Mou J, Pei DS. PAK5 promotes the migration and invasion of cervical cancer cells by phosphorylating SATB1. Cell Death Differ. 2019;26(6):994–1006. doi:10.1038/s41418-018-0178-4
32. Alvarado S, Wyglinski J, Suderman M, Andrews SA, Szyf M. Methylated DNA binding domain protein 2 (MBD2) coordinately silences gene expression through activation of the microRNA hsa-mir-496 promoter in breast cancer cell line. PLoS One. 2013;8(10):e74009. doi:10.1371/journal.pone.0074009
33. Karpinski P, Pesz K, Sasiadek MM. Pan-cancer analysis reveals presence of pronounced DNA methylation drift in CpG island methylator phenotype clusters. Epigenomics. 2017;9(11):1341–1352. doi:10.2217/epi-2017-0070
34. Jara-Espejo M, Peres Line SR. DNA G-quadruplex stability, position and chromatin accessibility are associated with CpG island methylation. FEBS J. 2020;287:483–495.
35. Sina AA, Carrascosa LG, Trau M. DNA methylation-based point-of-care cancer detection: challenges and possibilities. Trends Mol Med. 2019;29:955–966. doi:10.1016/j.molmed.2019.05.014
36. Liu Z, Wang Z, Jia E, et al. Analysis of genome-wide in cell free DNA methylation: progress and prospect. Analyst. 2019;144(20):5912–5922. doi:10.1039/C9AN00935C
37. Stirzaker C, Song JZ, Ng W, et al. Methyl-CpG-binding protein MBD2 plays a key role in maintenance and spread of DNA methylation at CpG islands and shores in cancer. Oncogene. 2017;36(10):1328–1338. doi:10.1038/onc.2016.297
38. Spiess BM, Wilcock BP, Physick-Sheard PW. Eosinophilic granulomatous dacryoadenitis causing bilateral keratoconjunctivitis sicca in a horse. Equine Vet J. 1989;21(3):226–228. doi:10.1111/j.2042-3306.1989.tb02153.x
39. Sansom OJ, Berger J, Bishop SM, Hendrich B, Bird A, Clarke AR. Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nat Genet. 2003;34(2):145–147. doi:10.1038/ng1155
40. Kanai Y, Ushijima S, Nakanishi Y, Hirohashi S. Reduced mRNA expression of the DNA demethylase, MBD2, in human colorectal and stomach cancers. Biochem Biophys Res Commun. 1999;264(3):962–966. doi:10.1006/bbrc.1999.1613
41. Ye D, Luo H, Lai Z, et al. ClC-3 chloride channel proteins regulate the cell cycle by up-regulating cyclin D1-CDK4/6 through suppressing p21/p27 expression in nasopharyngeal carcinoma cells. Sci Rep. 2016;6:30276. doi:10.1038/srep30276
42. Radine C, Peters D, Reese A, et al. The RNA-binding protein RBM47 is a novel regulator of cell fate decisions by transcriptionally controlling the p53-p21-axis. Cell Death Differ. 2020;27:1274–1285. doi:10.1038/s41418-019-0414-6
43. Aiello NM, Kang Y. Context-dependent EMT programs in cancer metastasis. J Exp Med. 2019;216(5):1016–1026. doi:10.1084/jem.20181827
44. Fiori ME, Di Franco S, Villanova L, Bianca P, Stassi G, De Maria R. Cancer-associated fibroblasts as abettors of tumor progression at the crossroads of EMT and therapy resistance. Mol Cancer. 2019;18(1):70. doi:10.1186/s12943-019-0994-2
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.