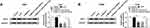Back to Journals » Cancer Management and Research » Volume 12
MALAT1 Promotes Cell Tumorigenicity Through Regulating miR-515-5p/EEF2 Axis in Non-Small Cell Lung Cancer
Authors Rong F, Liu L, Zou C, Zeng J, Xu Y
Received 15 December 2019
Accepted for publication 6 August 2020
Published 24 August 2020 Volume 2020:12 Pages 7691—7701
DOI https://doi.org/10.2147/CMAR.S242425
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Feng Rong, Liang Liu, Can Zou, Jing Zeng, Yasheng Xu
Department of Respiratory, Xiantao First People’s Hospital Affiliated to Changjiang University, Xiantao, Hubei, People’s Republic of China
Correspondence: Yasheng Xu Tel +86 728-3220705
Email [email protected]
Background: Previous studies suggested long noncoding RNA metastasis associated with lung adenocarcinoma transcript 1 (lncRNA MALAT1) acted as a tumor promoter to promote cell carcinogenesis in non-small cell lung cancer (NSCLC). MALAT1 was found to exist in serum exosomes of several cancers. However, the role of exosomal-derived MALAT1 in NSCLC remains poorly understood.
Materials and Methods: Exosomes were isolated using the ExoQuick precipitation kit. Western blot was used to detect the protein expression of CD3, CD63, apoptosis- and metastasis-related protein. The expression of MALAT1, microRNA (miR)-515-5p and eukaryotic elongation factor 2 (EEF2) mRNA was detected using quantitative real-time polymerase chain reaction. Cell viability, apoptosis, or invasion were measured using 3-(4, 5)-dimethylthiahiazo (-z-y1)-3, 5-di-phenytetrazoliumromide (MTT) assay, flow cytometry or transwell assay, respectively. The interaction between miR-515-5p and MALAT1 or EEF2 was confirmed by dual-luciferase reporter assay. In vivo experiments were conducted through the murine xenograft model.
Results: MALAT1 was highly expressed in serum and cell exosomes from NSCLC patients. MALAT1 knockdown repressed cell proliferation, invasion and induced cell apoptosis in vitro as well as inhibited tumor growth in vivo in NSCLC. Subsequently, we confirmed that MALAT1 was a sponge of miR-515-5p, and EEF2 was a target of miR-515-5p. Furthermore, MALAT1 served as a sponge of miR-515-5p to regulate EEF2 expression in NSCLC cells. More importantly, MALAT1 deletion performed anti-tumor effects by interacting with miR-515-5p/EEF2 axis in vitro and in vivo in NSCLC.
Conclusion: MALAT1 knockdown repressed NSCLC tumorigenicity by inhibiting cell proliferation, invasion and promoting apoptosis through regulating miR-515-5p/EEF2, besides, MALAT1 was highly enriched in exosomes of NSCLC, suggesting a possible molecular-targeted therapy for NSCLC patients.
Keywords: MALAT1, miR-515-5p, EEF2, NSCLC, tumorigenicity, exosomes
Introduction
Non-small cell lung cancer (NSCLC), accounting for around 80–85% of all lung cancers, is the most common sub-type of lung cancer, and has a low five-year survival rate of approximately 15%1,2 Even though improvement in early detection and multimodal therapy, the overall survival rate of NSCLC remains undesirable because of the metastasis, recurrence, and poor understanding on NSCLC molecular pathogenesis.3,4 Therefore, further clarifications on molecular mechanisms of NSCLC pathogenesis may shed light on the development of effective therapeutic approaches for NSCLC therapy.
Long noncoding RNAs (lncRNAs) have been revealed to be vital regulators in the development and progression of tumors by involving in cell cycle, growth, differentiation, metastasis, apoptosis and drug resistance.5–7 LncRNA metastasis associated with lung adenocarcinoma transcript 1 (lncRNA MALAT1) is a well-characterized functional lncRNA and has been reported to be aberrantly expressed in numerous human cancers, besides, MALAT1 dysregulation is closely implicated in the dysregulation of physiological processes, thereby affecting cancer development, progression, and response to therapy.8,9 Exosomes are extracellular vesicles with diameters ranging from 40 to 100 nm and can be secreted by most cell types, including cancer cells, in physiological and pathological conditions.10 Exosomes released by tumor cells are considered as mediators of tumor communication, which transmit and exchange their diverse cargoes, such as lncRNAs, between tumor cells and extracellular microenvironment.11,12 Exosomal lncRNAs have been identified to participate in cancer cell tumorigenesis and are potential clinical biomarkers for diagnosis and therapy of cancers.13,14 Recent studies have documented that MALAT1 exists in serum exosomes of several cancers, and plays important regulatory effects on tumor cell development and progression.15,16 Additionally, previous findings indicated MALAT1 was up-regulated and acted as an oncogene to promote cell carcinogenesis in NSCLC.17,18 However, the relationship between exosomal-derived MALAT1 and the development of NSCLC is still poorly understood.
It is well known that lncRNAs can function as sponges of miRNAs to affect the expression of miRNAs and the competition between mRNAs.19 MicroRNA-515-5p is a well-identified microRNA (miRNA) and has been reported to perform anti-tumor effects on the progression of several cancers.20 More importantly, Li et al recently revealed miR-515-5p suppressed survival and metastasis in NSCLC cells by interacting with CXCL6,21 indicating the potential therapeutic roles in NSCLC. Eukaryotic elongation factor 2 (EEF2) is a protein that induces GTP-dependent translocation of the ribosome and is important for protein synthesis. Besides that, it was also revealed that EEF2 had certain clinical values for the metastasis, diagnosis and prognosis.22,23 Therefore, it is necessary for us to better understand the roles of miR-515-5p and EEF2 in the pathogenesis of NSCLC.
In this study, we aimed to detect whether MALAT1 existed in serum and cell exosomes of NSCLC, investigate the molecular mechanism of MALAT1 in NSCLC cell proliferation, invasion and apoptosis, and explore the regulatory effects of MALAT1, miR-515-5p and EEF2 on the pathogenesis of NSCLC.
Materials and Methods
Serum Collection
Blood samples were collected from 52 patients with NSCLC and 52 healthy controls at Xiantao First People’s Hospital Affiliated to Changjiang University. All blood samples were centrifuged at 3000 g for 10 min after collecting for 1 h, and the supernatant serum was collected using RNase-free tubes. All subjects signed written informed consents and the Ethics Committee of Xiantao First People’s Hospital Affiliated to Changjiang University supported this study (with approval No. 20190304).
Cell Culture
Human Bronchial Epithelial cell Line 1 (HBE1) and human lung cancer epithelial cell lines (A549 and H1299) were purchased from Shanghai Academy of Life Science (Shanghai, China) and grown in the Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) harboring with 10% fetal bovine serum and 1% penicillin/streptomycin (Gibco) at 37°C with 5% CO2.
Exosome (Exo) Isolation
Serum exosomes were isolated using ExoQuick precipitation kit (System Biosciences, Mountain View, CA, USA) following the standard procedure. Four-milliliter serum sample was mixed with 1 mL ExoQuick solution and incubated for 30 min at 4°C, followed by centrifuging at 1500 g for 30 min. After removing the supernatant, exosomes were centrifuged again at 1500 g for 5 min to remove residual liquid.
Cell culture fluid from exosome-depleted medium was centrifuged at 3000 g for 30 min at 4°C to remove cellular debris/dead cells. Next, the resulting supernatant was further centrifuged at 100,000 g for 70 min at 4°C. After washing with PBS and further centrifuged at 100,000 g for 70 min, cell exosomes were obtained.
Transmission Electron Microscopy (TEM)
Ten-milliliter exosomes pellet was placed on a carbon-coated copper grid and incubated for 5 min at 37°C, and then was immersed with 2% phosphotungstic acid solution for 1 min. After washing 3 times with PBS, the preparations were captured using a transmission electron microscope (JEOL, Akishima, Japan).
Western Blot Analysis
Both exosomes and cells were lysed using the radio-immunoprecipitation assay (RIPA) buffer (Beyotime, Shanghai, China). Cell lysates were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, shifted onto a polyvinylidene fluoride membrane, and blocked with 5% non-milk. Next, the membranes were incubated with primary antibodies against CD9 (1:5000, ab68418, Abcam, Cambridge, MA, USA), CD63 (1:2000, ab68418, Abcam), Matrix metallopeptidase 9 (MMP-9) (1:1000, ab38898, Abcam), Vimentin (1:5000, ab92547, Abcam), B-cell lymphoma-2 (Bcl-2) (1:1000, ab692, Abcam), BCL2-associated X protein (Bax) (1:1000, ab32503, Abcam), and glyceraldehyde 3-phosphate dehydrogenase (GADPH) (1:10,000, ab181602, Abcam), followed by interaction with secondary HRP-conjugated antibody (1:1000, ab9482, Abcam). Finally, signals were visualized with the chemiluminescence chromogenic substrate (Beyotime).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA from NSCLC cells and tumor tissues was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and total RNA from exosomes was isolated with the exoRNeasy Midi Kit (Qiagen, Valencia, CA, USA) following the standard procedure. Then, total RNA was reversely transcribed into complementary DNA (cDNA) using All-in-One™ Kit (FulenGen, Guangzhou, China). After that, qRT-PCR was performed by SYBR™ Green Master Mix (Qiagen). Relative transcription alterations were analyzed by 2−ΔΔCt method and normalized by GADPH or U6. The specific primer sequences were listed as follows: MALAT1: F, 5ʹ-TCTTAGAGGGTGGGCTTTTGTT-3ʹ and R, 5ʹ-CTGCATCTAGGCCATCATACTG-3ʹ; EEF2: F, 5ʹ-GAGCTCTCCGAGAACGACC-3ʹ and R, 5ʹ-TACAGTGCCCAGGACAGGAT −3ʹ, miR-515-5p: F, 5ʹ-TTCTCCAAAAGAAAGCACTTTCTG-3ʹ and R, 5ʹ-TGGTGTCGTGGAGTCG-3ʹ; GADPH: F 5ʹ-GAGAAACCTGCCAAGTATGATGAC-3ʹ and R 5ʹ-GGAGTTGCTGTTGAAGTCAC-3ʹ, U6: F, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and R, 5ʹ-ACGCTTCACGAATTTGCGT-3ʹ.
Cell Transfection
The miR-515-5p mimic (miR-515-5p) and its negative control (miR-NC), miR-515-5p inhibitor (anti-miR-515-5p) and its negative control inhibitor (anti-NC) were purchased from RIBOBIO (Guangzhou, China). The short hairpin RNA (shRNA) targeting MALAT1 (sh-MALAT1), shRNA scramble control (sh-NC), small interfering RNA (siRNA) targeting MALAT1 (si-MALAT1), siRNA negative control (si-NC), empty vector (pcDNA), pcDNA-MALAT1 overexpression vector (MALAT1), pcDNA-EEF2 overexpression vector (EEF2) were synthesized by Genepharma (Shanghai, China). The transfection of oligonucleotides was performed using LipofectamineTM 3000 transfection reagent (Invitrogen).
3-(4, 5)-Dimethylthiahiazo (-z-y1)-3, 5-Di-Phenytetrazoliumromide (MTT) Assay
Transfected cells were seeded on the wells of a 96-well plate overnight. Then, the MTT reagent (Sigma, St. Louis, MO, USA) fixed with fresh medium (200 µL) was added into per well. Subsequently, the supernatant was discarded at 4 h post-reaction and 200 µL dimethyl sulfoxide (DMSO; Sigma) was supplemented to dissolve the formed precipitation. The optical density (OD) at 490 nm was detected using a spectrophotometer.
Cell Apoptosis Assay
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (BD Biosciences, San Jose, CA, USA) was used to determine apoptotic cells following the standard protocol. Briefly, transfected cells were resuspended with binding buffer, followed by staining with 5 μL FITC annexin V and 10 μL PI. Finally, the apoptotic rate was measured by a flow cytometer.
Transwell Assay
Transfected cells in serum-free DMEM were planted in the top chambers, whose membranes were pre-coated with the matrigel (BD Biosciences). Then, the lower chambers were filled with 500 μL DMEM mixed with 10% FBS. After 24 h, cells on the lower face of the membranes were fixed and stained. Finally, invaded cells were counted with a microscope.
Dual-Luciferase Reporter Assay
The wild-type (WT) or mutant (MUT) MALAT1 or EEF2 3ʹ-UTR possessing miR-515-5p binding sequences were cloned into the pmiR-RB-Report (Promega, Shanghai, China), respectively. Afterwards, these constructed vectors were co-transfected into A549 and H1299 cells with miR-515-5p or miR-NC using LipofectamineTM 3000 (Invitrogen) for 48 h. Lastly, a dual luciferase assay kit (Promega) was applied to analyze the luciferase activity.
Xenograft Experiments in vivo
Five-week-old male BALB/c nude mice (N=12) were used to establish xenograft models following the guidelines permitted by the Animal Ethics Committee of Xiantao First People’s Hospital Affiliated to Changjiang University. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines. H1299 cells stably injected with lentivirus-sh-NC or lentivirus-sh-MALAT1 were subcutaneously inoculated into the flanks of the nude mice. The tumor size in mice was detected every 4 days. The mice were sacrificed after 27 days, and the tumor masses were weighted and harvested for further molecular analysis.
Statistical Analysis
Data from three independent experiments were expressed as the mean ± standard deviation (SD). Statistical analysis was performed by GraphPad Prism 7 software (GraphPad Inc., San Diego, CA, USA). Significant differences between different groups were analyzed using Student’s t-test or one-way analysis of variance (ANOVA). P<0.05 suggested statistically significant.
Results
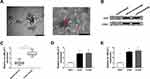
MALAT1 is Up-Regulated in Serum and Cell Exosomes from NSCLC Patients
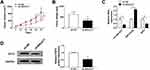
Exosomes from the serum of patients with NSCLC and healthy controls, as well as from human bronchial epithelial cell line 1 (HBE1) and human NSCLC cell lines (A549 and H1299) were extracted and characterized to explore the potential role of exosomes-derived MALAT1. TEM showed the morphology (round-shaped vesicles with double-layer membrane) and size (100 ± 60 nm) of exosomes (Figure 1A). Western blot analysis indicated the enrichment of CD9 and CD63 protein, the exosomal markers, in the exosomes-enriched fractions but not in exosomes-depleted supernatant (Figure 1B). These results indicated that exosomes were isolated from serum and cells successfully. Subsequently, the relative expression of MALAT1 in serum exosomes was detected and results showed MALAT1 was higher in serum exosomes from NSCLC patients than that from healthy controls (Figure 1C). Similarly, the level of MALAT1 was lower in cell exosomes from HBE1 compared with those from NSCLC cell lines (A549 and H1299) (Figure 1D). Moreover, we also observed that MALAT1 was elevated in NSCLC cell lines compared with the HBE1 (Figure 1E). All the results suggested that exosomal MALAT1 might be closely associated with the development of NSCLC.
Knockdown of MALAT1 Inhibits Cell Proliferation, Invasion and Induces Cell Apoptosis in NSCLC
Due to the up-regulation of MALAT1 in NSCLC cells and exosomes from NSCLC serum and cells, we explored the biological role of MALAT1 in vitro. Firstly, si-MALAT1 and si-NC were transfected into A549 and H1299 cells. As expected, MALAT1 was significantly decreased in A549 and H1299 cells transfected with si-MALAT1 (Figure 2A). After that, MTT assay showed knockdown of MALAT1 remarkably suppressed cell proliferation of A549 and H1299 cells (Figure 2B). Meanwhile, flow cytometric analysis implied that MALAT1 silence induced apoptosis of A549 and H1299 cells (Figure 2C). Additionally, MALAT1 deletion in A549 and H1299 cells markedly impaired cell invasion ability (Figure 2D). Furthermore, Western blot analysis showed a decreased expression of MMP9, Vimentin and Bcl-2 protein, but an increased level of Bax protein in A549 and H1299 cells (Figure 2E), further demonstrating MALAT1 deletion induced cell apoptosis and inhibited cell invasion in NSCLC. Taken together, MALAT1 silence inhibited cell tumorigenicity in NSCLC.
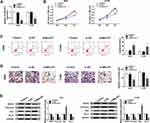
MALAT1 Silence Performs Anti-Tumor Effects by Binding to miR-515-5p in NSCLC Cells
It is well known that lncRNAs can function as sponges of miRNAs to affect the expression of miRNAs, thus affecting the development of cancers.7 Thus, we further investigated the potential miRNA targets of MALAT1. Using bioinformatics tool StarBase program, miR-515-5p was identified to contain the putative binding sites of MALAT1 (Figure 3A). Subsequently, a dual-luciferase reporter assay was conducted and declined luciferase activity in A549 and H1299 cells co-transfected with WT-MALAT1 and miR-515-5p confirmed the interaction between MALAT1 and miR-515-5p (Figure 3B). MiR-515-5p was found to be decreased in A549 and H1299 cells (Figure 3C). Besides that, after increasing the level of MALAT1 using MALAT1 overexpression vector in A549 and H1299 cells (Figure 3D), we discovered that MALAT1 up-regulation decreased miR-515-5p expression, while MALAT1 down-regulation increased miR-515-5p expression in A549 and H1299 cells (Figure 3E). These data indicated MALAT1 was a sponge of miR-515-5p and negatively regulated miR-515-5p expression in NSCLC cells.
Based on the relationship of MALAT1 and miR-515-5p, we hypothesized miR-515-5p might involve in MALAT1 silence-mediated inhibition on cell tumorigenicity. Firstly, A549 and H1299 cells were transfected with anti-miR-515-5p and anti-miR-NC, and a significant reduction of miR-515-5p expression was observed, indicating the successful transfection (Figure 3F). After that, si-NC, si-MALAT1, si-MALAT1 + anti-NC, or si-MALAT1 + anti-miR-515-5p was transfected into A549 and H1299 cells to perform rescue assay. Results showed miR-515-5p down-regulation could partially attenuate MALAT1 deletion-mediated inhibition on proliferation (Figure 3G and H), invasion (Figure 3J) and promotion on apoptosis (Figure 3I) of A549 and H1299 cells. Furthermore, the decrease of MMP9, Vimentin and Bcl-2 protein as well as the increase of Bax protein induced by MALAT1 deletion were also reversed by the inhibition of miR-515-5p in A549 and H1299 cells (Figure 3K and L). These results demonstrated that MALAT1 could interact with miR-515-5p to regulate cell carcinogenesis in NSCLC.
EEF2 is a Target of miR-515-5p and is Negatively Regulated by miR-515-5p
We further elucidated the molecular mechanisms underlying MALAT1/miR-515-5p mediated regulation on NSCLC progression. According to the prediction of StarBase program, miR-515-5p contained the binding sites of EEF2 (Figure 4A). Subsequently, the dual-luciferase reporter assay showed a great reduction of luciferase activity in A549 and H1299 cells co-transfected with WT-EEF2 and miR-515-5p, revealing the direct interaction between EEF2 and miR-515-5p (Figure 4B). EEF2 expression was increased in A549 and H1299 cells at mRNA and protein levels (Figure 4C and D). A549 and H1299 cells were transfected with miR-515-5p mimic or miR-NC, and an elevated expression of miR-515-5p was observed, indicating the successful transfection (Figure 4E). Immediately, we found miR-515-5p up-regulation decreased EEF2 expression, while miR-515-5p down-regulation increased EFE2 expression at mRNA and protein levels in A549 and H1299 cells (Figure 4F and G). Altogether, miR-515-5p targetedly suppressed EEF2 expression in NSCLC cells.
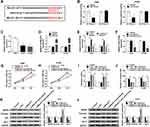
MiR-515-5p Inhibits Cell Proliferation, Invasion and Induces Cell Apoptosis in NSCLC by Targeting EEF2
Based on the miR-515-5p/EEF2 regulatory axis, we expounded the role of miR-515-5p/EEF2 axis in NSCLC progression. Firstly, A549 and H1299 cells were transfected with miR-NC, miR-515-5p, miR-515-5p + pcDNA, or miR-515-5p + EEF2, and we found EEF2 up-regulation reversed miR-515-5p restoration-mediated down-regulation of EEF2 expression in A549 and H1299 cells (Figure 5A and B). Next, rescue assay was performed, results displayed overexpressed miR-515-5p inhibited cell proliferation (Figure 5C), invasion (Figure 5E) and induced apoptosis (Figure 5D) in A549 and H1299 cells, while these effects could be partially overturned by EEF2 up-regulation (Figure 5C–E). Additionally, Western blot analysis also showed miR-515-5p up-regulation regulated invasion and apoptosis by reducing the expression of MMP9, Vimentin and Bcl-2 protein but increasing the level of Bax protein in A549 and H1299 cells, while these effects also were abated by EEF2 up-regulation (Figure 5F). Thus, we illustrated miR-515-5p functioned as a tumor suppressor to repress NSCLC progression by targeting EEF2.
MALAT1 May Participate in NSCLC Cell Tumorigenesis by Regulating EEF2 Through miR-515-5p
Based on the above results, we further analyzed the regulatory relationship among MALAT1, miR-515-5p and EEF2. Western blot analysis exhibited MALAT1 deletion reduced the expression of EEF2 in A549 cells, while this reduction could be rescued by miR-515-5p inhibition (Figure 6A). Moreover, the same changes were observed in H1299 cells (Figure 6B). Therefore, we proved that MALAT1 regulated EEF2 in a miR-515-5p-depended manner, and MALAT1 might regulate NSCLC cell tumorigenesis by regulating miR-515-5p/EEF2 axis.
MALAT1 Knockdown Inhibits NSCLC Tumor Growth in vivo
The carcinogenesis roles of MALAT1 in vivo were further elucidated, H1299 cells stably injected with lentivirus-sh-NC or lentivirus-sh-MALAT1 were subcutaneously inoculated into the flanks of the nude mice to establish xenograft models. Subsequently, we found MALAT1 knockdown inhibited NSCLC tumor growth, reflected by the inhibition of tumor volume and weight in sh-MALAT1 group (Figure 7A and B). After that, molecular analysis implied MALAT1 deletion reduced the levels of MALAT1 and EEF2, but increased the level of miR-515-5p in vivo (Figure 7C and D). Collectively, these results revealed that MALAT1 knockdown hindered NSCLC tumor growth in vivo through partially modulating miR-515-5p and EEF2 expression.
Discussion
Currently, although advances in early detection and multimodal therapy, the management of NSCLC is still unsatisfactory due to the poor therapeutic efficacy in defeating advanced cancer.24 Thus, ground-breaking therapeutic strategies need to be developed to prevent the progression and development of NSCLC. LncRNAs have been identified to play vital roles in cancer tumorigenesis and development, and are widely expressed in lung cancer.25 Additionally, because of the regulatory roles in diverse molecular pathways, lncRNAs are thought to be potential molecular-targeted therapeutic candidates for NSCLC.26 In recent, growing findings have revealed that exosomes contain specific lncRNAs, circRNAs and miRNAs and can protect them from degradation in the circulatory system, which are promising liquid biopsy for noninvasive cancer detection and therapy response monitoring.14,27
In this study, MALAT1 was highly expressed in serum and cell exosomes from NSCLC patients. Subsequently, we illustrated that MALAT1 was increased in NSCLC cells, and knockdown of MALAT1 prevented cell from proliferation, invasion and induced cell apoptosis in vitro as well as inhibited tumor growth in vivo in NSCLC. MALAT1 a well-known oncogene, and have been reported to have complex and extensive effects on the development and progression of tumors. For example, MALAT1 deletion alleviated the malignancy of anaplastic thyroid carcinoma by modulating FOXA1 via sponging miR-200a-3p.28 MALAT1 accelerated cell proliferation but attenuated cell apoptosis in ovarian cancer by binding to miR-503-5p.29 Consistently, MALAT1 knockdown restrained the metastasis of prostate cancer cells by directly interacting with miR-1-3p.30 Therefore, it is imperative to disclose exosomal-derived MALAT1 might be involved in the tumorigenesis of NSCLC cells.
MiRNAs are critical gene regulators and have been identified to function as oncogenic drivers and tumor suppressors in a variety of cancers, and are promising targets for novel anti-cancer therapies.31 It is well known that lncRNAs can function as sponges of miRNAs to regulate the gene expression through miRNAs, thereby involving in the development of cancers.7 The functional network between lncRNAs/miRNAs in cancer pathogenesis has attracted considerable attention. In this study, according to the bioinformatics analysis by StarBase program, MALAT1 was a sponge of miR-515-5p. Previous studies clarified that miR-515-5p was a crucial biomarker of cancers, and could act as a tumor suppressor to control cell multiple biological processes in prostate cancer,20 breast cancer,32 gastric cancer,33 and NSCLC.21 In the present study, miR-515-5p was found to be decreased and always performed anti-tumor roles by inhibiting proliferation, invasion and inducing apoptosis in NSCLC cells, which consistent with previous findings.
Furthermore, based on the interaction between MALAT1 and miR-515-5p, rescue assay was conducted and indicated MALAT1 deletion mediated inhibition on NSCLC cell tumorigenesis by binding to miR-515-5p. Besides that, this study also focused on the downstream target genes of miR-515-5p. We conformed that EEF2 was a target of miR-515-5p. What’s more, we discovered miR-515-5p functioned anti-proliferation, anti-invasion and pro-apoptosis effects by targeting EEF2 in NSCLC cells. Besides that, co-expression analysis suggested MALAT1 served as a sponge of miR-515-5p to regulate EEF2 expression in NSCLC cells. Thus, a MALAT1/miR-515-5p/EEF2 regulatory network was identified in NSCLC cells.
Conclusion
In general, MALAT1 deletion attenuated cell proliferation, invasion and accelerated cell apoptosis in vitro as well as inhibited tumor growth in vivo in NSCLC by regulating EEF2 through miR-515-5p, importantly, MALAT1 was high in the serum and cell exosomes of NSCLC patients, suggesting exosomal-derived MALAT1 might also reflect the biological changes occurring in NSCLC cells. All these results indicate MALAT1 may be a potential biomarker and have important significance for developing novel therapeutic strategies of NSCLC patients.
Author Contributions
All authors made substantial contribution to conception and design, acquisition of the data, or analysis and interpretation of the data; take part in drafting the article or revising it critically for important intellectual content; gave final approval of the revision to be published; and agree to be accountable for all aspect of the work.
Funding
No funding was received.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet. 2011;378(9804):1727–1740. doi:10.1016/S0140-6736(10)62101-0
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
3. Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382(9893):720–731. doi:10.1016/S0140-6736(13)61715-8
4. Simon J. Technology radiation technology targets tumors. Surgical precision without the incision. S D Med. 2014;67(9):362.
5. Ma Y, Zhang H, Li G, Hu J, Liu X, Lin L. LncRNA ANRIL promotes cell growth, migration and invasion of hepatocellular carcinoma cells via sponging miR-144. Anticancer Drugs. 2019;30(10):1013–1021. doi:10.1097/CAD.0000000000000807
6. Lin S, Zhang R, An X, et al. LncRNA HOXA-AS3 confers cisplatin resistance by interacting with HOXA3 in non-small-cell lung carcinoma cells. Oncogenesis. 2019;8(11):60. doi:10.1038/s41389-019-0170-y
7. Yu DJ, Zhong M, Wang WL. Long noncoding RNA CASC15 is upregulated in non-small cell lung cancer and facilitates cell proliferation and metastasis via targeting miR-130b-3p. Eur Rev Med Pharmacol Sci. 2019;23(18):7943–7949. doi:10.26355/eurrev_201909_19010
8. Eissmann M, Gutschner T, Hammerle M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–1087. doi:10.4161/rna.21089
9. Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, Zhu YS. MALAT1: a potential biomarker in cancer. Cancer Manag Res. 2018;10:6757–6768. doi:10.2147/CMAR.S169406
10. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364–372. doi:10.1016/j.tcb.2015.01.004
11. Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol. 2016;36(3):301–312. doi:10.1007/s10571-016-0366-z
12. Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13(10–11):1554–1571. doi:10.1002/pmic.201200329
13. Mohankumar S, Patel T. Extracellular vesicle long noncoding RNA as potential biomarkers of liver cancer. Brief Funct Genomics. 2016;15(3):249–256. doi:10.1093/bfgp/elv058
14. Zheng R, Du M, Wang X, et al. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer. 2018;17(1):143. doi:10.1186/s12943-018-0880-3
15. Qiu JJ, Lin XJ, Tang XY, Zheng TT, Lin YY, Hua KQ. Exosomal metastasis associated lung adenocarcinoma transcript 1 promotes angiogenesis and predicts poor prognosis in epithelial ovarian cancer. Int J Biol Sci. 2018;14(14):1960–1973. doi:10.7150/ijbs.28048
16. Zhang P, Zhou H, Lu K, Lu Y, Wang Y, Feng T. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. Onco Targets Ther. 2018;11:291–299. doi:10.2147/OTT.S155134
17. Wei S, Wang K, Huang X, Zhao Z, Zhao Z. LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis. Int J Immunopathol Pharmacol. 2019;33:2058738419859699. doi:10.1177/2058738419859699
18. Yang T, Li H, Chen T, Ren H, Shi P, Chen M. LncRNA MALAT1 depressed chemo-sensitivity of NSCLC cells through directly functioning on miR-197-3p/p120 catenin axis. Mol Cells. 2019;42(3):270–283. doi:10.14348/molcells.2019.2364
19. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
20. Zhang X, Zhou J, Xue D, Li Z, Liu Y, Dong L. MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer. Int J Biol Macromol. 2019;129:227–232. doi:10.1016/j.ijbiomac.2019.01.127
21. Li J, Tang Z, Wang H, et al. CXCL6 promotes non-small cell lung cancer cell survival and metastasis via down-regulation of miR-515-5p. Biomed Pharmacother. 2018;97:1182–1188. doi:10.1016/j.biopha.2017.11.004
22. Sun HG, Dong XJ, Lu T, Yang MF, Wang XM. Clinical value of eukaryotic elongation factor 2 (eEF2) in non-small cell lung cancer patients. Asian Pac J Cancer Prev. 2014;14(11):6533–6535. doi:10.7314/APJCP.2013.14.11.6533
23. Cheng D, He Z, Zheng L, Xie D, Dong S, Zhang P. PRMT7 contributes to the metastasis phenotype in human non-small-cell lung cancer cells possibly through the interaction with HSPA5 and EEF2. Onco Targets Ther. 2018;11:4869–4876. doi:10.2147/OTT.S166412
24. Lu T, Wang Y, Chen D, Liu J, Jiao W. Potential clinical application of lncRNAs in non-small cell lung cancer. Onco Targets Ther. 2018;11:8045–8052. doi:10.2147/OTT.S178431
25. Chi Y, Wang J, Wang J, Yu W, Yang J. Long Non-Coding RNA in the pathogenesis of cancers. Cells. 2019;8(9):9. doi:10.3390/cells8091015
26. Ricciuti B, Mencaroni C, Paglialunga L, et al. Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy. Med Oncol. 2016;33(2):18. doi:10.1007/s12032-016-0731-2
27. He M, Zeng Y. Microfluidic exosome analysis toward liquid biopsy for cancer. J Lab Autom. 2016;21(4):599–608. doi:10.1177/2211068216651035
28. Gou L, Zou H, Li B. Long noncoding RNA MALAT1 knockdown inhibits progression of anaplastic thyroid carcinoma by regulating miR-200a-3p/FOXA1. Cancer Biol Ther. 2019;20(11):1355–1365. doi:10.1080/15384047.2019.1617567
29. Sun Q, Li Q, Xie F. <p>LncRNA-MALAT1 regulates proliferation and apoptosis of ovarian cancer cells by targeting miR-503-5p. Onco Targets Ther. 2019;12:6297–6307. doi:10.2147/OTT.S214689
30. Dai X, Liang Z, Liu L, Guo K, Xu S, Wang H. Silencing of MALAT1 inhibits migration and invasion by sponging miR13p in prostate cancer cells. Mol Med Rep. 2019;20(4):3499–3508. doi:10.3892/mmr.2019.10602
31. Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15(5):429. doi:10.2174/138920101505140828161335
32. Pinho FG, Frampton AE, Nunes J, et al. Downregulation of microRNA-515-5p by the estrogen receptor modulates sphingosine kinase 1 and breast cancer cell proliferation. Cancer Res. 2013;73(19):5936–5948. doi:10.1158/0008-5472.CAN-13-0158
33. Zhang H, Wang X, Huang H, Wang Y, Zhang F, Wang S. Hsa_circ_0067997 promotes the progression of gastric cancer by inhibition of miR-515-5p and activation of X chromosome-linked inhibitor of apoptosis (XIAP). Artif Cells Nanomed Biotechnol. 2019;47(1):308–318. doi:10.1080/21691401.2018.1553787
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.