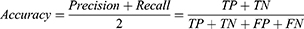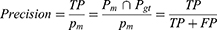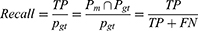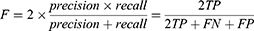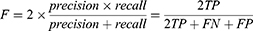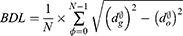Back to Journals » Clinical Ophthalmology » Volume 16
Machine Learning and Deep Learning Techniques for Optic Disc and Cup Segmentation – A Review
Authors Alawad M, Aljouie A , Alamri S, Alghamdi M , Alabdulkader B , Alkanhal N, Almazroa A
Received 9 November 2021
Accepted for publication 11 February 2022
Published 11 March 2022 Volume 2022:16 Pages 747—764
DOI https://doi.org/10.2147/OPTH.S348479
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Mohammed Alawad,1 Abdulrhman Aljouie,1 Suhailah Alamri,2,3 Mansour Alghamdi,4 Balsam Alabdulkader,4 Norah Alkanhal,2 Ahmed Almazroa2
1Department of Biostatistics and Bioinformatics, King Abdullah International Medical Research Center, King Saud bin Abdulaziz University for Health Sciences, Riyadh, Saudi Arabia; 2Department of Imaging Research, King Abdullah International Medical Research Center, King Saud bin Abdulaziz University for health Sciences, Riyadh, Saudi Arabia; 3Research Labs, National Center for Artificial Intelligence, Riyadh, Saudi Arabia; 4Department of Optometry and Vision Sciences College of Applied Medical Sciences, King Saud University, Riyadh, Saudi Arabia
Correspondence: Ahmed Almazroa; Abdulrhman Aljouie, Email [email protected]; [email protected]
Background: Globally, glaucoma is the second leading cause of blindness. Detecting glaucoma in the early stages is essential to avoid disease complications, which lead to blindness. Thus, computer-aided diagnosis systems are powerful tools to overcome the shortage of glaucoma screening programs.
Methods: A systematic search of public databases, including PubMed, Google Scholar, and other sources, was performed to identify relevant studies to overview the publicly available fundus image datasets used to train, validate, and test machine learning and deep learning methods. Additionally, existing machine learning and deep learning methods for optic cup and disc segmentation were surveyed and critically reviewed.
Results: Eight fundus images datasets were publicly available with 15,445 images labeled with glaucoma or non-glaucoma, and manually annotated optic disc and cup boundaries were found. Five metrics were identified for evaluating the developed models. Finally, three main deep learning architectural designs were commonly used for optic disc and optic cup segmentation.
Conclusion: We provided future research directions to formulate robust optic cup and disc segmentation systems. Deep learning can be utilized in clinical settings for this task. However, many challenges need to be addressed before using this strategy in clinical trials. Finally, two deep learning architectural designs have been widely adopted, such as U-net and its variants.
Keywords: glaucoma, glaucoma screening, fundus images, big images data
Introduction
Glaucoma is an eye disease that damages the optic nerve and causes visual impairment (7.7 million).1 According to the World Health Organization (WHO), an estimated 2.2 billion people worldwide have a vision impairment, including 1 billion classified as moderate to severe. Various reports have been published on glaucoma prevalence worldwide (Table 1).
 |
Table 1 Summary of Glaucoma Prevalence Studies Around the World |
Glaucoma is known as the “silent theft” of sight because the associated vision loss results from increasing ocular pressure on the optic nerve, which is often asymptomatic. Thus, early diagnosis of glaucoma is essential to prevent irreversible vision loss. One major factor in the early detection of glaucoma is identifying optic nerve damage, which can be detected by using a fundus camera developed for this purpose.14 Fundus imaging is a non-invasive procedure that relies on the monofocal indirect ophthalmoscopy principle.
Computer vision problems have significantly benefitted from recent advancements in deep learning methods. Image segmentation is a fundamental component of computer vision and visual-understanding problems. It can be defined as the sub-division of an image or video based on a distinct visual region with semantic meaning. Over the past few years, deep learning has achieved greater accuracy rates than traditional image processing techniques and has performed well by many popular segmentation benchmarks. This has led to a paradigm shift in image segmentation.15,16
Therefore, the paradigm shift towards deep learning in image segmentation has affected biomedical imaging applications. Currently, deep learning has become an efficient and effective methodology for a wide variety of biomedical image segmentation tasks in cardiac imaging,17 neuroimaging,18,19 and abdominal radiology.20
To date, segmentation with deep learning has been used to detect, localize, and diagnose clinical problems such as breast cancer,21 lung cancer,22 and Alzheimer’s Disease.23 Deep learning segmentation has been successfully used with most major imaging modalities, including magnetic resonance imaging (MRI),24 computed tomography (CT),25 and ultrasound.26
The use of computer-aided diagnosis (CAD) systems in ophthalmology to segment the required features has expanded over the last two decades, particularly using fundus images or optical coherence tomography (OCT) for glaucoma and retinal diseases such as diabetic retinopathy. This expanding interest has led scientists to investigate machine learning and deep learning techniques for CAD systems in recent years, which has generated a large volume of published methodologies, methods, and datasets. Thus, review studies can play a crucial role in advancing CAD systems by providing specialists with a summary of the current landscape and pathways for future investigation. Previous review studies have examined artificial intelligence (AI) and its applications in ophthalmology, both in general and in relation to specific conditions, including glaucoma.27–30
This review is structured as follows: a comprehensive literature review is performed in the following section. After that, the material and methods section is introduced. In the fourth section, the results are presented. Finally, the fifth and sixth sections constitute the discussion and conclusion.
Literature Review
It is noteworthy that reviews describing the general use of AI in ophthalmology tend to target a broad audience, leading to limited technical conclusions with implications for specific ocular conditions. Ting et al30 presented a thorough review of deep learning techniques for ophthalmology, highlighting technical and clinical domains. They provided, in detail, conventional deep learning models, training, datasets, reference standards (ground truths), and outcome measures. Furthermore, this review discussed many ocular conditions such as diabetic retinopathy, retrolental fibroplasia (RLF), age-related macular degeneration (AMD), and glaucoma. Sengupta et al31 discussed and compared the most important deep learning techniques with applications in ophthalmology, specifically focusing on glaucoma.
Owing to the increasing prevalence of glaucoma, many AI methods, image processing techniques, and glaucoma-related datasets have been published in recent years, along with several glaucoma-specific reviews.32–45 Mursch-Edlmayr et al35 presented a clinically focused review exploring AI-based strategies for detecting and monitoring glaucoma. This review examined various glaucoma testing modalities, such as optical coherence tomography (OCT), visual field (VF) testing, and fundus photos, with minimal technical discussion. They concluded that AI-based algorithms for analyzing fundus images have high translational potential due to the accessibility and simplicity of fundus photography. Future integration of AI-based analysis of fundus images may help address the current limitations of glaucoma management around the world.
Several technical review studies have also been published examining machine learning approaches and network architectures employed for glaucoma assessment and management. Hagiwara et al33 reviewed CAD systems for glaucoma diagnosis based on conventional machine learning. They examined the entire CAD procedure, including pre-processing techniques, segmentation methods, feature extraction strategies, feature selection approaches, and classification methods. They concluded that integrating deep learning methods simplifies these systems and improves their reliability. Thakur and Juneja32 compared recent segmentation and classification approaches for glaucoma diagnosis from fundus images; they also discussed the limitations of these methods and possible avenues to increase their efficiency. Barros et al34 reviewed machine learning approaches for diagnosing and detecting glaucoma. Their review examined supervised methods for glaucoma prediction from fundus images and categorized them into deep learning and non-deep learning approaches. Eswari and Karkuzhali’s46 summarised the advantages and disadvantages of many segmentation and classification techniques.
This study examines and critiques the deep learning methods that have been explicitly designed for optic cup and disc segmentation to diagnose glaucoma from fundus images. This review will be considering the methods section covering the most public fundus images datasets, the evaluation metrics used to evaluate the developed deep learning segmentation approaches, and finally, the most developed machine learning and deep learning architectures for optic disc and optic cup segmentation.
Materials and Methods
Research Criteria
The required information was retrieved for glaucoma (optic disc and optic cup boundaries) machine learning and deep learning segmentation approaches. The image datasets presented in this review were obtained from various websites, conference papers, and journal peer review studies. Therefore, this review was not required to obtain ethical approval to be conducted.
Study Selection
Standard academic web search engines were used, such as Google Scholar, PubMed, and Web of Science. Glaucoma, optic disc, and cup segmentation were placed after or before the following keywords or phrases: deep learning, machine learning, artificial intelligence, telemedicine, fundus datasets, and fundus databases. The search was conducted from October 2020 to March 2021.
Results
Datasets
Multiple fundus retinal image datasets were developed for glaucoma detection. Several datasets were also developed for different retinal diseases, such as diabetic retinopathy, which this review will not consider. These images may not meet crucial criteria, such as sufficient image quality, which can subsequently limit the efficacy of machine learning analyses. In addition, these image sets may be insufficient for the development of machine learning processes for certain diseases that focus on specific areas of the retina, as is the case with the optic nerve head in glaucoma. Moreover, some datasets, such as RIM-ONE and Drishti-GS, only have a small number of images, hindering the development and testing of machine learning algorithms. Therefore, efforts have been made to develop task-oriented datasets to overcome the limitations of existing datasets and identify the images required to train algorithms to detect glaucoma automatically. This study reviews the available retinal images datasets currently used for automated glaucoma detection (Table 2).
 |
Table 2 Current Available Public Datasets with Labelling and Manual Annotation for Glaucoma Fundus Images |
Evaluation Metrics
Evaluating segmentation maps for optic disc and cup segmentation is not a straightforward task. In the literature, various evaluation metrics have been used to assess the performances of machine learning and deep learning models. The optic cup and disc are objects in segmentation tasks where both the ground truth and segmentation encompass both foreground and background partitions. The evaluation of a segmentation algorithm can focus on assessing how well the pixels of the ground truth are detected; therefore, most researchers use a pixel-wise approach to assess the efficacy of their models. To further assess the quality of the detected object, some researchers have used the boundary assessment approach.
In this section, we will discuss the evaluation metrics used in the literature. First, we need to define the relevant notation. Given a segmentation method m and a ground truth gt, denote Pm and Nm as the positive and negative pixels of an image, respectively, such that Im = Pm  Nm is the resulting object detected. Similarly, the ground truth is represented as I_gt = Pgt
Nm is the resulting object detected. Similarly, the ground truth is represented as I_gt = Pgt  Ngt, where Pgt and Ngt are the positive and negative pixels, respectively. To achieve optimal detection, a model should aim for Pm= Pgt. For a pixel-based object detection measurement, there are four fundamental indicators as follows:
Ngt, where Pgt and Ngt are the positive and negative pixels, respectively. To achieve optimal detection, a model should aim for Pm= Pgt. For a pixel-based object detection measurement, there are four fundamental indicators as follows:
- True positive (TP) results represent the pixels that are labelled and predicted as an object, such that TP = Pm ∩ Pgt.
- False negative (FN) results represent the pixels that are labelled as an object but predicted as non-object, such that FN = Nm ∩ Pgt.
- False positive (FP) results represent the pixels that are not labelled as an object but are predicted an object, such that FP = Pm ∩ Ngt.
- True negative (TN) results represent the pixels that are both labelled and predicted as non-object, such that TN = Nm ∩ Ngt.
Pixel Accuracy
Pixel accuracy56 reflects the ratio of successful object detection using the total number of true positives and true negatives to report the total number of correct predictions. This metric is a standard and simple evaluation technique that was initially developed for classification. However, pixel accuracy is used similarly for segmentation evaluation by reporting the percentage of pixels in the image that are correctly classified. The mathematical expression for pixel accuracy is as follows:
Generally, pixel accuracy is a misleading metric when the object representation within the image is small because the measure will be biased in reporting how well the negative case is identified. This bias is very relevant in evaluating the accuracy of optic cup detection in a fundus image, because this structure is typically a small portion of the fundus image.
Precision and Recall
Two of the most popular evaluation metrics for both classification and segmentation are precision and recall.57 Precision, also known as specificity, evaluates how accurate the model is predicting in positive pixels; whereas recall, also known as sensitivity, measures the percentage of correctly identified true positives TP. Precision and recall can be expressed mathematically from the pixel-wise perspective as follows:
When the cost of an FP is very high and the cost of an FN is low, precision is the recommended evaluation metric. However, when the cost of an FN is high, recall is the more important metric. Generally, precision and recall are reported together in evaluating optic cup and disc segmentation.
Dice (F-Measure)
The trade-off between precision and recall can be estimated using the F-measure, commonly known as DICE, which can effectively combine precision and recall into one formula58 and has since become a popular evaluation technique for many segmentation problems. In fact, DICE has been used as the official evaluation metric for the widely known Retinal Fundus Glaucoma Challenge Edition (REFUGE) challenge.48 F-measure can be expressed as
Jaccard Index
The Jaccard index,59 also known as the intersection of union (IoU), is one of the most common evaluation metrics in image segmentation. This coefficient is defined as the size of the intersection divided by the size of the union of the sample sets:
Both the Jaccard index and DICE provide similar results; in other words, if one metric says that Model A offers better segmentation than Model B, the other metric will produce the same result. However, there are some minor differences between the metrics, such as the penalisation of single instances of misclassifications. Because this study focuses on the general concepts of these metrics, we refer the reader to the following studies for more details.60,61
Boundary Distance Localisation
One way to evaluate segmentation methods is to consider a boundary approach instead of a pixel-based approach. A pixel-based approach measures how well the pixels of the ground truth are detected while a boundary-based approach focuses on how accurately the boundaries are represented.60 The boundary approach can better reflect the quality of object detection than the pixel-based approach in some object detection problems, such as optic disc segmentation. A common boundary-based evaluation metric is the mean boundary distance in pixels between the predicted segmentation results and the ground truth. This measure is known as boundary distance localisation (BDL). BDL can be defined mathematically as
where 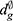 and
and 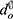 are the distance from the center of the cup or disc to the predicted and ground truth boundaries, respectively. This evaluation method was used by Jiang et al62 and produced high marginal variability for disc segmentation when compared to the traditional pixel-based metric, such as F-measure.
are the distance from the center of the cup or disc to the predicted and ground truth boundaries, respectively. This evaluation method was used by Jiang et al62 and produced high marginal variability for disc segmentation when compared to the traditional pixel-based metric, such as F-measure.
An objective assessment of different proposed methods for OC and OD segmentation is presented in Table 3 using mean dice, sensitivity, specificity, and Jaccard on the REFUGE test set. In the table, the comparison shows that the state-of-the-art graph convolutional network based (GCN)63 achieved a mean dice coefficient of 95.58% and 97.76% for OD and OC, respectively.
 |
Table 3 Comparison of Optic Cup (OC) and Optic Disc (OD) Segmentation Methods on REFUGE Test Set Using Different Metrics |
Machine Learning and Deep Learning Architectures
The abundance of newly generated datasets coupled with growing computing power has led to diverse deep learning approaches to image analysis. Image segmentation has benefited from this advancement, revolutionizing traditional image processing techniques.65 More deep learning approaches have begun to appear specifically for segmentation because the nature of segmentation tasks have expanded to applications for autonomous vehicles,66 augmented reality, video surveillance, and medical imaging. This has improved the advancement of deep learning.
As an important task in medical image analysis, fundus image segmentation has become a significant contributor to the evolution of deep learning methods. In this section, we will review different deep learning architectures that have been employed for optic cup and disc segmentation (Table 4). We divide these methodologies into four major categories: convolutional neural network-related approaches, U-Net-related methods, generative adversarial network (GAN) approaches, and other deep learning-related approaches.
 |
Table 4 Deep Learning and Machine Learning Methods for Optic Cup and Disc Segmentation |
A Convolutional Neural Network (CNN)
CNN is a type of feed-forward artificial neural network (ANN). CNN’s comprise stacked layers of convolution and pooling operations, followed by batch normalization layers to speed up learning and stabilize the input of subsequent layers as the network grows deeper. In the final layers, weight vectors are concatenated and passed into one or more fully connected layers for classification output. The convolution layers slide over the input with a fixed step and window size to control the moving dot product of the data. The flattened output is then passed into a differentiable nonlinear activation function, typically a rectified linear unit (ReLU). As a result of its success in computer vision tasks over state-of-the-art conventional machine learning algorithms, such as support vector machines, many researchers have investigated the ability of CNNs to segment the optic cup and disc regions,93–95 and retinal blood vessels96 in fundus images. Unlike image-level classification or bounding box-level prediction tasks, the feature maps in semantic segmentation are resized to the original image space. Each pixel is given a probability of being assigned to a given semantic label.
Tan et al94 used a CNN with two convolutions followed by a max-pooling layer that connects to a fully connected layer with 100 neurons and a final layer of 4 neurons as an output to classify each pixel of a fundus image as either background, optic cup, fovea, or blood vessel. Figure 2 shows the authors’ proposed CNN architecture (Figure 2). Sreng et al93 employed DeepLab-v3+,97 which comprises an encoder and decoder module, for optic disc segmentation. The network design uses spatial pyramid pooling (Figure 1), accounting for different optic disc sizes across fundus images with varying scales. The authors built and evaluated their optic disc semantic segmentation models with 2787 retinal images from five different publicly available datasets.
 |
Figure 1 Tan et al proposed CNN architecture. Notes: Reprinted from: Tan JH, Acharya UR, Bhandary SV, Chua KC, Sivaprasad S. Segmentation of optic disc, fovea and retinal vasculature using a single convolutional neural network. J Comput Sci. 2017;20:70–79.94 Copyright 2017, with permission from Elsevier. |
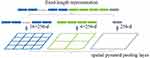 |
Figure 2 Spatial pyramid pooling layer: pooling features extracted using different window sizes on the feature maps. Notes: © 2015 IEEE. Reprinted, with permission, from: He K, Zhang X, Ren S, Sun J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans Pattern Anal Mach Intell. 2015;37(9):1904–1916.98. |
Sun et al82 reframed optic disc segmentation as an object detection problem because the shape of the optic disc is a non-rotated ellipse. They used Faster R-CNN,99 with VGG-16 as a backbone for the R-CNN.
Lu et al100 explored weakly supervised learning. This approach aimed to stochastically cluster every pixel through pairwise pixel information into the background and foreground for optic disc segmentation using two fully connected layers. The network was simultaneously trained on three different tasks: 1) optic disc segmentation, 2) glaucoma classification, and 3) evidence map (heatmap for affected regions) prediction.
Shankaranarayana et al101 proposed a pipeline for an end-to-end, fully connected convolutional encoder-decoder network with two paths for the encoder component of the network. This pipeline took two modalities as input: an RGB fundus image and a depth map. The authors used a pre-trained network to predict the depth map for monocular retinal depth estimation. Subsequently, the original image and the predicted depth map were fed into another encoder-decoder network for optic cup and disc segmentation.
U-Net Based Approaches
Unlike other machine learning and deep learning methodologies, U-Net, a fully convolutional network (FCN) sub-type, was explicitly developed for biomedical imaging segmentation.102 The architecture and training strategy of a U-Net-based approach promotes the efficient use of annotated data for more reliable prediction. The architectural design is a compression path that consists of consecutive convolutional layers and a max-pooling layer to extract features while limiting the feature map size (Figure 3: U-Net Architecture). Skip connections, an alternative path for backpropagation, were used to share localization information and expand layers.
 |
Figure 3 U-net architecture (example for 32×32 pixels in the lowest resolution). Each blue box corresponds to a multi-channel feature map. The number of channels is denoted on top of the box. The x-y-size is provided at the lower left edge of the box. White boxes represent copied feature maps. The arrows denote the different operation. Notes: Reprinted by permission from Springer Nature from: Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A (eds). Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham; 2015:234–241.102 Copyright © Springer International Publishing Switzerland 2015. Available from: https://link.springer.com/book/10.1007/978-3-319-24574-4. |
This unique architecture captured attention after winning multiple segmentation challenges in 2015 and has since been widely used, especially for medical applications. Most neural network-based fundus segmentation methods for glaucoma have relied on U-Net or U-Net alternatives. Sevastopolsky81 modified U-Net and used the Jaccard Index and DICE score to compare their methods with two different CNN-based approaches.76,103
Chakravarty and Sivaswamy70 proposed a glaucoma assessment framework that jointly segmented and classified fundus images. They proposed a multi-task CNN architecture wherein U-Net is used for segmentation, and the output and latent layers are used together to predict glaucoma. Al-Bander et al72 proposed DenseNet, which simultaneously segments the optic cup and disc. DenseNet resembles the traditional U-Net architecture with some differences, such as a Dense Block, transition Down Block, and transition Up Block. They presented a comprehensive comparison using multiple datasets and evaluation metrics to compare their method with deep learning-based and traditional image processing techniques.
Fu et al73 presented the Disc Aware Ensemble Network (DENet) for glaucoma screening that incorporates a global fundus image and an image that focuses on the optic disc region. However, as the DENet was used for glaucoma screening, the U-Net network was used for segmenting the disc to influence the screening decision. As such, their method was evaluated only for glaucoma screening rather than segmentation performance. Fu et al78 also introduced a novel joint optic cup and disc segmentation approach based on a polar transformation of the fundus image. To this end, they proposed M-Net, which is a multi-scale U-Net with a side-output layer that produces a probability map.
Sevastopolsky et al87 proposed a U-Net-based, special cascade network that incorporated the concept of iterative refinement. The model was designed as stacked multiple blocks to achieve better recognition where each block was a basic U-Net. They compared their model with one U-Net-based model81 and two CNN-based approaches.76,103
Wang et al68 introduced a coarse-to-fine deep learning model based on U-Net designed specifically to segment the optic disc region. The coarse-to-fine strategy splits the segmentation task into multiple stages: one for extracted vessels (vessel density map) and the other for local disc patches to obtain segmentation results. The authors compared the performance of their disc segmentation method against several deep learning and image processing methods on various datasets. Although this method is novel from a prepossessing perspective, it did not consistently improve across all datasets and did not include optic cup segmentation.
Gu et al71 proposed a context encoder network (CE-NET) for medical image segmentation. CE-NET modified the U-Net architecture to account for the consecutive pooling and striated convolution operation, leading to spatial information loss. They replaced the U-Net encoder block with a ResNet-34 pre-trained block and added a context extractor that consists of a dense atrous convolution block and a residual multi-kernel pooling block. This method was proposed for general medical image segmentation and was evaluated for optic disc segmentation. The model was evaluated using the standard pixel-based mean and standard deviation.
Finally, Yu et al85 proposed a robust optic cup and disc segmentation method by modifying the U-Net architecture. To this end, they adopted the ResNet-34 pre-trained block for encoding and kept the original U-Net block for decoding. Krishna Adithya et al114 developed “EffUnet”, a two-phase network to detect glaucoma by first segmenting cup and disc using efficient Unet and then predicting glaucoma using a generative adversarial network.
Generative Adversarial Network Approaches
Deep learning architecture focused on decision-making features such as classification, regression, or segmentation in the past. However, generative networks such as variational autoencoders104 and adversarial networks introduced a creative component to neural networks. A generative adversarial network (GAN) framework was introduced by Goodfellow et al105 in 2014. GAN attracted attention owing to the simplicity of the network structure (Figure 4) and its robust generative performance. GAN essentially consisted of two main components: a generator that captured data distribution and a discriminator that estimated the probability of a certain feature. One primary application of GAN was an image-to-image translation, which is defined as translating one possible representation of a scene to another. This concept allowed GAN to be used for segmentation by decoding the input image into the required label. Many researchers have used this concept to solve major segmentation problems, such as semantic segmentation106 or specific medical image segmentation problems.107 Accordingly, many researchers have used this approach to segment optic cups and discs to improve the performance of current methods.
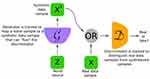 |
Figure 4 GAN general architecture consists of Generator (G) which output a synthetic sample given a noise variable input and a Discriminator (D) which estimate the probability of a given sample coming from real dataset. Both components are built based on neural network. Notes: © 2018 IEEE. Reprinted, with permission, from: Creswell A, White T, Dumoulin V, Arulkumaran K, Sengupta B, Bharath AA. Generative adversarial networks: An overview. IEEE Signal Process Mag. 2018;35(1):53–65.108. |
Singh et al84 adopted a conditional GAN109 with a generative deep learning network that learned invariant features to segment the optic disc. A major component of the network, according to the authors, improved the segmentation performance in the skip connection to the encoder-decoder within the generator network.
Liu et al79 introduced a semi-supervised segmentation model based on a conditional GAN. Their model consisted of a segmentation net and a generator that increased the training set while the discriminator identified fake images to ensure compatible training. This method demonstrated the advantages of semi-supervised methods by utilizing unlabelled data, which could help mitigate the lack of labeled data. The segmentation performance model was compared to traditional U-Net, M-Net,78 and other architectures. Wang et al83 used a patch-based output space adversarial learning framework that jointly segmented the optic cup and disc. Their model exploited unsupervised domain adaptation to address the domain shift across datasets that usually reduced the performance of the segmentation model when it was trained on a specific dataset and tested on a different dataset. They also introduced a morphology-aware segmentation tool that produced better segmentation loss.
Son et al89 introduced another GAN-based framework for retinal vessel and optic disc segmentation alternative to CNN-based methods. Their model showed significant performance improvement for retinal vessel segmentation and no improvement for optic disc segmentation. Jing et al62 proposed GL-NET, a hybrid deep-CNN and GAN model for segmenting the optic cup and disc. The generator was a full CNN that included an encoder-decoder to extract features using the VGG16 network.110 They added skip connections within the generator to promote low- and high-level feature information. To better address the variability between models, they adopted the boundary approach BDL and pixel-based evaluation metrics to compare the model’s performance.
Building a unified model that generalizes across image types was challenging due to the large variety of fundus cameras. Wang et al111 addressed this issue by creating a domain-invariant model based on a GAN. Their model adopted domain adaptation methods built on source domain data and utilized a small target dataset to improve the performance in the target domain. The model used the DICE coefficient to compare the performance with other significant studies. Bisneto et al75 proposed a full glaucoma detection system that utilized GAN for image segmentation. They built an entire system to detect glaucoma, their segmentation method focused on the optic disc only, and performance was evaluated accordingly.
Other Deep Learning-Related Architectures
The three approaches described above are the most common in machine learning and deep learning for glaucoma-related image segmentation. However, some researchers have investigated different strategies and architectures and have achieved exciting results. Sedai et al88 proposed a cascaded shape regression network that learned the final shape for segmentation. They employed a boosted regression tree and proposed a data augmentation approach to improve segmentation performance. Their evaluation used the DRISHTI dataset. Zhang et al74 presented embedded edge-attention representations to guide a segmentation network called ET-Net. Their model followed an encoder-decoder with two additional components for guidance and aggregation. The model was tested on multiple segmentation tasks, including optic disc and cup segmentation.
Ding et al77 introduced hierarchical attention networks for medical image segmentation. The approach combined encoder-decoder networks with CNNs to extract feature maps. Three blocks were added, specifically a dense similarity block, an attention propagation block, and an information aggregation block. The model was tested on the REFUGE dataset. Tabassum et al67 introduced an encoder-decoder network (CDED-Net) for optic cup and disc segmentation. This architecture was slightly different from the typical U-Net because there was no bottleneck layer, and fewer convolutional layers were employed.
Discussion
This review outlines the abundant deep-learning-based optic cup and disc segmentation models from fundus images. This technique has enormous potential for clinical application because it is a robust method that overcomes many of the challenges associated with traditional image processing techniques.32 Researchers widely use three main network architectures for accomplishing the optic cup and disc segmentation task; importantly, each network architecture has its advantages and disadvantages. The traditional CNN offers excellent optic cup and disc segmentation; however, it typically requires many accurately labeled images to reach its full potential. The U-Net architecture is the current state-of-the-art architecture owing to its training simplicity and data efficiency. However, its multi-scale skip connection tends to use unnecessary information, and low-level encoder features are insufficient, leading to poorer performance. The GAN method uses a creative image-image translation component to achieve accurate optic cup and disc segmentation; however, current GAN methods are deterministic, making them difficult to generalize. Finally, further investigation of generative stochastic models is needed to study the randomness effect because the natural world is stochastic.
There are some common challenges with methods for optic disc and cup segmentation from fundus images that need to be addressed to translate these tools to clinical settings:
● Reliable and broadly applicable deep learning models in computer vision commonly rely on a large set of training and testing data. However, as observed in the reviewed models, these publicly available datasets are often relatively small. Accordingly, accumulating a large set of data labeled by specialists is the most critical challenge in building a reliable and generalizable model. Although researchers have attempted to account for this issue using strategies such as transfer learning, the need for a large volume of high-quality data for large-scale evaluation of segmentation methods remains.
● Current fundus image annotation of the optic cup and disc is subjectively performed by ophthalmologists, inducing the inter– and intra-observer variability. Almazroa et al112 studied this phenomenon in-depth. They concluded that the variability in annotation results from the unclear optic disc and cup boundaries are human-related factors such as examiner fatigue or lack of concentration, or image-related factors such as low quality, hazy, unfocused images, or display devices that are too dim or too bright. This variability poses a challenge for scientists building a segmentation model using a non-standardized labeling mechanism. To account for this problem, an effective segmentation model must address these differences while training and learning unambiguous weights to perform better than human identification ultimately.
● There are a variety of fundus imaging modalities that acquire the images of the eye’s posterior pole with different resolutions, angles, and degrees of the posterior segment. Furthermore, some imaging modalities require dilated pupils, whereas others require non-dilated pupils. This creates a domain shift problem, a well-known and well-defined problem in machine learning. Most of the segmentation models reviewed in this study utilize training and test data with the same image features, ignoring the significant real-life challenge posed by inter-model variability. Wang and Deng113 discussed various techniques to address this problem, essential for building a reliable automated diagnostic tool.
● Evaluating a segmentation model is not as straightforward as evaluating classification. It is difficult to compare and identify the best architecture for optic cup and disc segmentation. This challenge created variabilities among researchers who used different metrics (reviewed in section Evaluation Metrics) to assess their work. A unified evaluation criterion for segmentation models is critical to compare different studies effectively. Such standardized evaluation criteria should include an evaluation metric that considers optic cups, disc vulnerabilities, and difficulties and uses a common evaluation methodology. To this end, we suggest that boundary approaches may be more valuable in assessing the quality of detected objects than pixel-based methods.
Devising an efficient but low-performing degradation model for OC and OD segmentation (by reducing the model complexity) is of high importance114,115 for the enhancement of both training and inference times. Additionally, developing an explainable model is another topic that many believe is crucial for Glaucoma AI-based detection tools to be approved and accepted in the clinic.28,116
The future direction for better optic cup and disc segmentation should account for the above cases. A large set of annotated data should be published to create and evaluate better models. Because the annotation is not a straightforward task, public data must consider inter and intra-observer variability by providing multi annotations per image. More advanced machine learning methodologies must be experimented with to overcome image discrepancies to achieve a fully robust model. Lastly, a unified evaluation metric must be adopted for upcoming models. One pixel-based approach such as F-score and a boundary approach such as boundary distance localization are recommended.
Clinical Value of Automated Optic Disc and Optic Cup Segmentation
The current advancement in artificial intelligence within healthcare focuses on enhancing various aspects such as improving accessibility, increasing speed, reducing cost, early diagnosis, and enhancing human abilities. The Singapore Integrated Diabetic Retinopathy program (SiDRP) is a telemedicine-based screening program for diabetic retinopathy.117 The SiDRP started to integrate AI algorithms into their workflow with the ultimate goal of fully automating the screening process, increasing efficiency, and reducing the cost for the whole program.118
One proven application for AI within healthcare is clinical decision support. Many clinical diagnoses and screening, especially medical imaging, are time-consuming and labour-intensive. Thus, AI offers an appealing solution to expedite decision-making and improve efficiency. Various companies began to offer an AI-based clinical support system for chest x-ray screening, brain MRI, breast mammography, and diabetic retinopathy.
Glaucoma diagnosis is a complex problem requiring well-trained ophthalmologists or optometrists. Unfortunately, there is a limited number of trained specialists worldwide to examine all suspected cases. A glaucoma decision support system could offer an alternative route to this problem. Such a system could expedite the specialist’s decision-making and may eventually evolve as an automated screening system. The Cup and disc segmentation system is a major milestone and fundamental to reaching the ultimate destination. Once a system is mature enough, it will revolutionize the clinical practice to screen and diagnose glaucoma.
As been mentioned previously, cup and disc segmentation undergo an inter-observer variability. Such disagreements are related to various factors; some are human-related factors. As with the nature of humans, examiners might experience fatigue (marking at the last hour) and lack of concentration (marking in a rush or during busy hours). It has been recommended that a second opinion could improve the overall segmentation.112 However, a second opinion might not be available all the time due to various reasons. Therefore, an AI-based segmentation system could be that second opinion once mature.
Conclusion
Effective optic cup and disc segmentation on fundus images are critical for accurate automated diagnosis and prediction of glaucoma. Machine learning and deep learning models have shown promising results for various segmentation tasks, both for fundus image analysis and other applications. This study presents a comprehensive review of the current deep learning and machine learning models explicitly designed for optic cup and disc segmentation on fundus images. We also reviewed the available retinal image datasets and common evaluation metrics to provide a broad understanding of how automated image segmentation tools are designed, tested, and evaluated. While three network architectures have been commonly used for optic cup and disc segmentation, the U-net architecture and the GAN model have demonstrated robust results. They may have the potential to be tested in clinical settings shortly. However, many challenges still need to be addressed to create robust image segmentation models that can be applied in clinical settings or large-scale diagnosis campaigns.
Disclosure
The authors report no conflicts of interest in this work.
References
1. World Health Organization (WHO). Blindness and vision impairment (2021). Available from: https://www.who.int/news-room/fact-sheets/detail/blindness-and-visual-impairment.
2. Quigley HA. Number of people with glaucoma worldwide. Br J Ophthalmol. 1996;80(5):389–393. doi:10.1136/bjo.80.5.389
3. Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90(3):262–267. doi:10.1136/bjo.2005.081224
4. Eid TM, el-Hawary I, el-Menawy W. Prevalence of glaucoma types and legal blindness from glaucoma in the western region of Saudi Arabia: a hospital-based study. Int Ophthalmol. 2009;29(6):477–483. doi:10.1007/s10792-008-9269-4
5. Al obeidan SA, Dewedar A, Osman EA, Mousa A. The profile of glaucoma in a Tertiary Ophthalmic University Center in Riyadh, Saudi Arabia. Saudi J Ophthalmol. 2011;25(4):373–379. doi:10.1016/j.sjopt.2011.09.001
6. Day AC, Baio G, Gazzard G, et al. The prevalence of primary angle closure glaucoma in European derived populations: a systematic review. Br J Ophthalmol. 2012;96(9):1162–1167. doi:10.1136/bjophthalmol-2011-301189
7. Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121(11):2081–2090. doi:10.1016/j.ophtha.2014.05.013
8. Kapetanakis VV, Chan MP, Foster PJ, Cook DG, Owen CG, Rudnicka AR. Global variations and time trends in the prevalence of primary open angle glaucoma (POAG): a systematic review and meta-analysis. Br J Ophthalmol. 2016;100(1):86–93. doi:10.1136/bjophthalmol-2015-307223
9. Chan EWE, Li X, Tham YC, et al. Glaucoma in Asia: regional prevalence variations and future projections. Br J Ophthalmol. 2016;100(1):78–85. doi:10.1136/bjophthalmol-2014-306102
10. Gupta P, Zhao D, Guallar E, Ko F, Boland MV, Friedman DS. Prevalence of glaucoma in the United States: the 2005–2008 national health and nutrition examination survey. Invest Ophthalmol Vis Sci. 2016;57(6):2905–2913. doi:10.1167/iovs.15-18469
11. Flaxman SR, Bourne RR, Resnikoff S, et al. Global causes of blindness and distance vision impairment 1990–2020: a systematic review and meta-analysis. Lancet Glob Health. 2017;5(12):e1221–e1234. doi:10.1016/S2214-109X(17)30393-5
12. Khandekar R, Chauhan D, Yasir ZH, Al-Zobidi M, Judaibi R, Edward DP. The prevalence and determinants of glaucoma among 40 years and older Saudi residents in the Riyadh Governorate (except the Capital)–A community based survey. Saudi J Ophthalmol. 2019;33(4):332–337. doi:10.1016/j.sjopt.2019.02.006
13. Zhang N, Wang J, Chen B, Li Y, Jiang B. Prevalence of primary angle closure glaucoma in the last 20 years: a meta-analysis and systematic review. Front Med. 2020;7:624179.
14. Myers JS, Fudemberg SJ, Lee D. Evolution of optic nerve photography for glaucoma screening: a review. Clin Experiment Ophthalmol. 2018;46(2):169–176. doi:10.1111/ceo.13138
15. Minaee S, Boykov YY, Porikli F, Plaza AJ, Kehtarnavaz N, Terzopoulos D. Image segmentation using deep learning: a survey. IEEE Trans Pattern Anal Mach Intell. 2021;1. doi:10.1109/TPAMI.2021.3059968
16. Ghosh S, Das N, Das I, Maulik U. Understanding deep learning techniques for image segmentation. ACM Comput Surv. 2019;52(4):1–35. doi:10.1145/3329784
17. Chen C, Qin C, Qiu H, et al. Deep learning for cardiac image segmentation: a review. Front Cardiovasc Med. 2020;7:25. doi:10.3389/fcvm.2020.00025
18. Akkus A, Galimzianova Z, Hoogi A, Rubin DL, Erickson BJ. Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging. 2017;30(4):449–459. doi:10.1007/s10278-017-9983-4
19. Işın A, Direkoğlu C, Şah M. Review of MRI-based brain tumor image segmentation using deep learning methods. Procedia Comput Sci. 2016;102:317–324. doi:10.1016/j.procs.2016.09.407
20. Rehman A, Khan FG. A deep learning based review on abdominal images. In: Multimedia Tools and Applications; 2020:1–32.
21. Krithiga R, Geetha P. Breast cancer detection, segmentation and classification on histopathology images analysis: a systematic review. Arch Comput Methods Eng. 2021;28(4):2607–2619. doi:10.1007/s11831-020-09470-w
22. Li Z, Zhang J, Tan T, et al. Deep learning methods for lung cancer segmentation in whole-slide histopathology images-the acdc@ lunghp challenge 2019. IEEE J Biomed Health Inform. 2020;25(2):429–440.
23. Yamanakkanavar N, Choi JY, Lee B. MRI segmentation and classification of human brain using deep learning for diagnosis of Alzheimer’s disease: a survey. Sensors. 2020;20(11):3243. doi:10.3390/s20113243
24. Liu J, Pan Y, Li M, et al. Applications of deep learning to MRI images: a survey. Big Data Min Anal. 2018;1(1):1–18.
25. Domingues I, Pereira G, Martins P, Duarte H, Santos J, Abreu PH. Using deep learning techniques in medical imaging: a systematic review of applications on CT and PET. Artif Intell Rev. 2020;53(6):4093–4160. doi:10.1007/s10462-019-09788-3
26. Akkus Z, Cai J, Boonrod A, et al. A survey of deep-learning applications in ultrasound: artificial intelligence–powered ultrasound for improving clinical workflow. J Am Coll Radiol. 2019;16(9):1318–1328. doi:10.1016/j.jacr.2019.06.004
27. Balyen L, Peto T. Promising artificial intelligence-machine learning-deep learning algorithms in ophthalmology. Asia Pac J Ophthalmol. 2019;8(3):264–272. doi:10.22608/APO.2018479
28. Ting DSW, Pasquale LR, Peng L, et al. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol. 2019;103(2):167–175. doi:10.1136/bjophthalmol-2018-313173
29. Thompson AC, Jammal AA, Medeiros FA. A review of deep learning for screening, diagnosis, and detection of glaucoma progression. Transl Vis Sci Technol. 2020;9(2):42. doi:10.1167/tvst.9.2.42
30. Ting DS, Peng L, Varadarajan AV, et al. Deep learning in ophthalmology: the technical and clinical considerations. Prog Retin Eye Res. 2019;72:100759. doi:10.1016/j.preteyeres.2019.04.003
31. Sengupta S, Singh A, Leopold HA, Gulati T, Lakshminarayanan V. Ophthalmic diagnosis using deep learning with fundus images–A critical review. Artif Intell Med. 2020;102:101758. doi:10.1016/j.artmed.2019.101758
32. Thakur N, Juneja M. Survey on segmentation and classification approaches of optic cup and optic disc for diagnosis of glaucoma. Biomed Signal Process Control. 2018;42:162–189. doi:10.1016/j.bspc.2018.01.014
33. Hagiwara Y, Koh JEW, Tan JH, et al. Computer-aided diagnosis of glaucoma using fundus images: a review. Comput Methods Programs Biomed. 2018;165:1–12. doi:10.1016/j.cmpb.2018.07.012
34. Barros DM, Moura JC, Freire CR, Taleb AC, Valentim RA, Morais PS. Machine learning applied to retinal image processing for glaucoma detection: review and perspective. Biomed Eng Online. 2020;19(1):1–21. doi:10.1186/s12938-020-00767-2
35. Mursch-Edlmayr AS, Ng WS, Diniz-Filho A, et al. Artificial intelligence algorithms to diagnose glaucoma and detect glaucoma progression: translation to clinical practice. Transl Vis Sci Technol. 2020;9(2):55. doi:10.1167/tvst.9.2.55
36. Almazroa A, Burman R, Raahemifar K, Lakshminarayanan V. Optic disc and optic cup segmentation methodologies for glaucoma image detection: a survey. J Ophthalmol. 2015;2015. doi:10.1155/2015/180972
37. Almazroa A, Sun W, Alodhayb S, Raahemifar K, Lakshminarayanan V. Optic disc segmentation for glaucoma screening system using fundus images. Clin Ophthalmol. 2017;11:2017–2029. doi:10.2147/OPTH.S140061
38. Almazroa A, Alodhayb S, Raahemifar K, Lakshminarayanan V. An automatic image processing system for glaucoma screening. Int J Biomed Imaging. 2017;2017. doi:10.1155/2017/4826385
39. Almazroa A, Alodhayb S, Raahemifar K, Lakshminarayanan V. Optic cup segmentation: type-II fuzzy thresholding approach and blood vessel extraction. Clin Ophthalmol. 2017;11:841. doi:10.2147/OPTH.S117157
40. Gopalakrishnan A, Almazroa A, Raahemifar K, Lakshminarayanan V. Optic disc segmentation using circular Hough transform and curve fitting. In:
41. Biran A, Bidari PS, Almazroa A, Lakshminarayanan V, Raahemifar K. Blood vessels extraction from retinal images using combined 2D Gabor wavelet transform with local entropy thresholding and alternative sequential filter. In:
42. Almazroa A, Alodhayb S, Burman R, Sun W, Raahemifar K, Lakshminarayanan V. Optic cup segmentation based on extracting blood vessel kinks and cup thresholding using Type-II fuzzy approach. In:
43. Burman R, Almazroa A, Raahemifar K, Lakshminarayanan V. Automated detection of optic disc in fundus images. In: Advances in Optical Science and Engineering. New Delhi: Springer; 2015:327–334.
44. Almazroa A, Sun W, Alodhayb S, Raahemifar K, Lakshminarayanan V. Optic disc segmentation: level set methods and blood vessels inpainting. In: Medical Imaging 2017: Imaging Informatics for Healthcare, Research, and Applications. Vol. 10138. International Society for Optics and Photonics; March, 2017:1013806.
45. Almazroa AA, Woodward MA, Newman-Casey PA, et al. The appropriateness of digital diabetic retinopathy screening images for a computer-aided glaucoma screening system. Clin Ophthalmol. 2020;14:3881. doi:10.2147/OPTH.S273659
46. Eswari MS, Karkuzhali S. Survey on segmentation and classification methods for diagnosis of glaucoma. In:
47. Fumero F, Alayón S, Sanchez JL, Sigut J, Gonzalez-Hernandez M. RIM-ONE: an open retinal image database for optic nerve evaluation. In:
48. Orlando JI, Fu H, Breda JB, et al. Refuge challenge: a unified framework for evaluating automated methods for glaucoma assessment from fundus photographs. Med Image Anal. 2020;59:101570. doi:10.1016/j.media.2019.101570
49. Sivaswamy J, Krishnadas SR, Joshi GD, Jain M, Tabish AUS. Drishti-gs: retinal image dataset for optic nerve head (onh) segmentation. In:
50. Carmona EJ, Rincón M, García-Feijoó J, Martínez-de-la-casa JM. Identification of the optic nerve head with genetic algorithms. Artif Intell Med. 2008;43(3):243–259. doi:10.1016/j.artmed.2008.04.005
51. Cheng J, Zhang Z, Tao D, et al. Similarity regularized sparse group lasso for cup to disc ratio computation. Biomed Opt Express. 2017;8(8):3763–3777. doi:10.1364/BOE.8.003763
52. Zhang Z, Yin FS, Liu J, et al. Origa-light: an online retinal fundus image database for glaucoma analysis and research. In
53. Almazroa A, Alodhayb S, Osman E, et al. Retinal fundus images for glaucoma analysis: the Riga dataset. In: Medical Imaging 2018: Imaging Informatics for Healthcare, Research, and Applications. Vol. 10579. International Society for Optics and Photonics; March, 2018:105790B.
54. Li L, Xu M, Liu H, et al. A large-scale database and a CNN model for attention-based glaucoma detection. IEEE Trans Med Imaging. 2019;39(2):413–424. doi:10.1109/TMI.2019.2927226
55. Diaz-Pinto A, Morales S, Naranjo V, et al. CNNs for automatic glaucoma assessment using fundus images: an extensive validation. Biomed Eng Online. 2019;18(1):1–19. doi:10.1186/s12938-019-0649-y
56. Menditto A, Patriarca M, Magnusson B. Understanding the meaning of accuracy, trueness and precision. Accreditation Qual Assur. 2007;12(1):45–47. doi:10.1007/s00769-006-0191-z
57. Powers DM. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. J Mach Learn Technol. 2011;2(1):37-63. doi:10.48550/arXiv.2010.16061
58. Dice LR. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;26:297-302. doi:10.2307/1932409.
59. Jaccard P. The distribution of the flora in the alpine zone. New Phytologist. 1912;11:37-50. doi:10.1111/j.1469-8137.1912.tb05611.x
60. Pont-Tuset J, Marques F. Supervised evaluation of image segmentation and object proposal techniques. IEEE Trans Pattern Anal Mach Intell. 2015;38(7):1465–1478. doi:10.1109/TPAMI.2015.2481406
61. Margolin R, Zelnik-Manor L, Tal A. How to evaluate foreground maps?
62. Jiang Y, Tan N, Peng T. Optic disc and cup segmentation based on deep convolutional generative adversarial networks. IEEE Access. 2019;7:64483–64493. doi:10.1109/ACCESS.2019.2917508
63. Tian Z, Zheng Y, Li X, Du S, Xu X. Graph convolutional network based optic disc and cup segmentation on fundus images. Biomed Opt Express. 2020;11(6):3043–3057. doi:10.1364/BOE.390056
64. Liu B, Pan D, Song H. Joint optic disc and cup segmentation based on densely connected depthwise separable convolution deep network. BMC Med Imaging. 2021;21(1):1–12. doi:10.1186/s12880-020-00528-6
65. Kuruvilla J, Sukumaran D, Sankar A, Joy SP. A review on image processing and image segmentation. In:
66. Treml M, Arjona-Medina J, Unterthiner T, et al. Speeding up semantic segmentation for autonomous driving; 2016.
67. Tabassum M, Khan TM, Arsalan M, et al. CDED-Net: joint segmentation of optic disc and optic cup for glaucoma screening. IEEE Access. 2020;8:102733–102747. doi:10.1109/ACCESS.2020.2998635
68. Wang L, Liu H, Lu Y, Chen H, Zhang J, Pu J. A coarse-to-fine deep learning framework for optic disc segmentation in fundus images. Biomed Signal Process Control. 2019;51:82–89. doi:10.1016/j.bspc.2019.01.022
69. Kälviäinen RVJPH, Uusitalo H. DIARETDB1 diabetic retinopathy database and evaluation protocol. In: Medical Image Understanding and Analysis. Vol. 2007. Citeseer; 2007:61.
70. Chakravarty A, Sivswamy J. A deep learning based joint segmentation and classification framework for glaucoma assessment in retinal color fundus images; 2018. Available from: https://arxiv.org/abs/1808.01355. Accessed February 26, 2022.
71. Gu Z, Cheng J, Fu H, et al. Ce-net: context encoder network for 2d medical image segmentation. IEEE Trans Med Imaging. 2019;38(10):2281–2292. doi:10.1109/TMI.2019.2903562
72. Al-Bander B, Williams BM, Al-Nuaimy W, Al-Taee MA, Pratt H, Zheng Y. Dense fully convolutional segmentation of the optic disc and cup in colour fundus for glaucoma diagnosis. Symmetry. 2018;10(4):87. doi:10.3390/sym10040087
73. Fu H, Cheng J, Xu Y, et al. Disc-aware ensemble network for glaucoma screening from fundus image. IEEE Trans Med Imaging. 2018;37(11):2493–2501. doi:10.1109/TMI.2018.2837012
74. Zhang Z, Fu H, Dai H, Shen J, Pang Y, Shao L. Et-net: a generic edge-attention guidance network for medical image segmentation. In:
75. Bisneto TRV, de Carvalho Filho AO, Magalhães DMV. Generative adversarial network and texture features applied to automatic glaucoma detection. Appl Soft Comput. 2020;90:106165. doi:10.1016/j.asoc.2020.106165
76. Zilly J, Buhmann JM, Mahapatra D. Glaucoma detection using entropy sampling and ensemble learning for automatic optic cup and disc segmentation. Comput Med Imaging Graph. 2017;55:28–41. doi:10.1016/j.compmedimag.2016.07.012
77. Ding F, Yang G, Liu J, et al. Hierarchical attention networks for medical image segmentation; 2019. Available from: https://arxiv.org/abs/1911.08777. Accessed February 26, 2022.
78. Fu H, Cheng J, Xu Y, Wong DWK, Liu J, Cao X. Joint optic disc and cup segmentation based on multi-label deep network and polar transformation. IEEE Trans Med Imaging. 2018;37(7):1597–1605. doi:10.1109/TMI.2018.2791488
79. Liu S, Hong J, Lu X, et al. Joint optic disc and cup segmentation using semi-supervised conditional GANs. Comput Biol Med. 2019;115:103485. doi:10.1016/j.compbiomed.2019.103485
80. Kim M, Han JC, Hyun SH, et al. Medinoid: computer-aided diagnosis and localization of glaucoma using deep learning. Appl Sci. 2019;9(15):3064. doi:10.3390/app9153064
81. Sevastopolsky A. Optic disc and cup segmentation methods for glaucoma detection with modification of U-Net convolutional neural network. Pattern Recognit Image Anal. 2017;27(3):618–624. doi:10.1134/S1054661817030269
82. Sun X, Xu Y, Zhao W, You T, Liu J. Optic disc segmentation from retinal fundus images via deep object detection networks. In:
83. Wang S, Yu L, Yang X, Fu CW, Heng PA. Patch-based output space adversarial learning for joint optic disc and cup segmentation. IEEE Trans Med Imaging. 2019;38(11):2485–2495. doi:10.1109/TMI.2019.2899910
84. Singh VK, Rashwan HA, Akram F, et al. Retinal optic disc segmentation using conditional generative adversarial network. In: CCIA; October, 2018:373–380.
85. Yu S, Xiao D, Frost S, Kanagasingam Y. Robust optic disc and cup segmentation with deep learning for glaucoma detection. Comput Med Imaging Graph. 2019;74:61–71. doi:10.1016/j.compmedimag.2019.02.005
86. Joshua AO, Nelwamondo FV, Mabuza-Hocquet G. Segmentation of optic cup and disc for diagnosis of glaucoma on retinal fundus images. In:
87. Sevastopolsky A, Drapak S, Kiselev K, Snyder BM, Keenan JD, Georgievskaya A. Stack-u-net: refinement network for improved optic disc and cup image segmentation. In: Medical Imaging 2019: Image Processing. Vol. 10949. International Society for Optics and Photonics; March, 2019:1094928.
88. Sedai S, Roy P, Mahapatra D, Garnavi R. Segmentation of optic disc and optic cup in retinal fundus images using coupled shape regression.
89. Son J, Park SJ, Jung KH. Towards accurate segmentation of retinal vessels and the optic disc in fundoscopic images with generative adversarial networks. J Digit Imaging. 2019;32(3):499–512. doi:10.1007/s10278-018-0126-3
90. Bajwa MN, Malik MI, Siddiqui SA, et al. Two-stage framework for optic disc localization and glaucoma classification in retinal fundus images using deep learning. BMC Med Inform Decis Mak. 2019;19(1):1–16. doi:10.1186/s12911-018-0723-6
91. Budai A, Odstrcilik J, Kolar R, et al. A public database for the evaluation of fundus image segmentation algorithms. Invest Ophthalmol Vis Sci. 2011;52(14):1345.
92. Zhao R, Liao W, Zou B, Chen Z, Li S. Weakly-supervised simultaneous evidence identification and segmentation for automated glaucoma diagnosis. In:
93. Sreng S, Maneerat N, Hamamoto K, Win KY. Deep learning for optic disc segmentation and glaucoma diagnosis on retinal images. Appl Sci. 2020;10(14):4916. doi:10.3390/app10144916
94. Tan JH, Acharya UR, Bhandary SV, Chua KC, Sivaprasad S. Segmentation of optic disc, fovea and retinal vasculature using a single convolutional neural network. J Comput Sci. 2017;20:70–79. doi:10.1016/j.jocs.2017.02.006
95. Zilly JG, Buhmann JM, Mahapatra D. Boosting convolutional filters with entropy sampling for optic cup and disc image segmentation from fundus images. In: International Workshop on Machine Learning in Medical Imaging. Cham: Springer; October, 2015:136–143.
96. Yao Z, Zhang Z, Xu LQ. Convolutional neural network for retinal blood vessel segmentation. In:
97. Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In:
98. He K, Zhang X, Ren S, Sun J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans Pattern Anal Mach Intell. 2015;37(9):1904–1916. doi:10.1109/TPAMI.2015.2389824
99. Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. Adv Neural Inf Process Syst. 2015;28:91–99.
100. Lu Z, Chen D, Xue D, Zhang S. Weakly supervised semantic segmentation for optic disc of fundus image. J Electron Imaging. 2019;28(3):033012.
101. Shankaranarayana S, Ram SM, Mitra K, Sivaprakasam M. Fully convolutional networks for monocular retinal depth estimation and optic disc-cup segmentation. IEEE J Biomed Health Inform. 2019;23(4):1417–1426. doi:10.1109/JBHI.2019.2899403
102. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In:
103. Maninis KK, Pont-Tuset J, Arbeláez P, Van Gool L. Deep retinal image understanding. In:
104. Kingma DP, Welling M. Auto-encoding variational Bayes; 2013. Available from: https://arxiv.org/abs/1312.6114. Accessed February 26, 2022.
105. Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial networks. Commun ACM. 2020;63(11):139–144. doi:10.1145/3422622
106. Luc P, Couprie C, Chintala S, Verbeek J. Semantic segmentation using adversarial networks; 2016. Available from: https://arxiv.org/abs/1611.08408. Accessed February 26, 2022.
107. Frid-Adar M, Diamant I, Klang E, Amitai M, Goldberger J, Greenspan H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing. 2018;321:321–331. doi:10.1016/j.neucom.2018.09.013
108. Creswell A, White T, Dumoulin V, Arulkumaran K, Sengupta B, Bharath AA. Generative adversarial networks: an overview. IEEE Signal Process Mag. 2018;35(1):53–65. doi:10.1109/MSP.2017.2765202
109. Isola P, Zhu JY, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. In:
110. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition; 2014. Available from: https://arxiv.org/abs/1409.1556. Accessed February 26, 2022.
111. Wang S, Yu L, Heng PA. Optic disc and cup segmentation with output space domain adaptation. Refug Rep. 2019. Available from: http://rumc-gcorg-p-public.s3.amazonaws.com/f/challenge/229/431a27fd-58e3-4719-88d7-d59929a4e8b1/REFUGE-CUHKMED.pdf. Accessed February 26, 2022.
112. Almazroa A, Alodhayb S, Osman E, et al. Agreement among ophthalmologists in marking the optic disc and optic cup in fundus images. Int Ophthalmol. 2017;37(3):701–717. doi:10.1007/s10792-016-0329-x
113. Wang M, Deng W. Deep visual domain adaptation: a survey. Neurocomputing. 2018;312:135–153. doi:10.1016/j.neucom.2018.05.083
114. Krishna Adithya V, Williams BM, Czanner S, et al. EffUnet-SpaGen: an efficient and spatial generative approach to glaucoma detection. J Imaging. 2021;7(6):92. doi:10.3390/jimaging7060092
115. Jin B, Liu P, Wang P, Shi L, Zhao J. Optic disc segmentation using attention-based U-Net and the improved cross-entropy convolutional neural network. Entropy. 2020;22(8):844. doi:10.3390/e22080844
116. Yousefi S, Pasquale LR, Boland MV. Artificial intelligence and glaucoma: illuminating the black box. Ophthalmol Glaucoma. 2020;3(5):311–313. doi:10.1016/j.ogla.2020.04.008
117. Nguyen HV, Tan GS, Tapp RJ, et al. Cost-effectiveness of a national telemedicine diabetic retinopathy screening program in Singapore. Ophthalmology. 2016;123(12):2571–2580. doi:10.1016/j.ophtha.2016.08.021
118. Grzybowski A, Brona P, Lim G, et al. Artificial intelligence for diabetic retinopathy screening: a review. Eye. 2020;34(3):451–460. doi:10.1038/s41433-019-0566-0
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.