Back to Journals » Infection and Drug Resistance » Volume 14
Machine Learning and Artificial Intelligence for the Prediction of Host–Pathogen Interactions: A Viral Case
Authors Yakimovich A
Received 29 May 2021
Accepted for publication 3 August 2021
Published 20 August 2021 Volume 2021:14 Pages 3319—3326
DOI https://doi.org/10.2147/IDR.S292743
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Artur Yakimovich
Artificial Intelligence for Life Sciences CIC, London, UK
Correspondence: Artur Yakimovich
Artificial Intelligence for Life Sciences CIC, 9 High Street, London, N8 7FJ, UK
Email [email protected]
Abstract: The research of interactions between the pathogens and their hosts is key for understanding the biology of infection. Commencing on the level of individual molecules, these interactions define the behavior of infectious agents and the outcomes they elicit. Discovery of host–pathogen interactions (HPIs) conventionally involves a stepwise laborious research process. Yet, amid the global pandemic the urge for rapid discovery acceleration through the novel computational methodologies has become ever so poignant. This review explores the challenges of HPI discovery and investigates the efforts currently undertaken to apply the latest machine learning (ML) and artificial intelligence (AI) methodologies to this field. This includes applications to molecular and genetic data, as well as image and language data. Furthermore, a number of breakthroughs, obstacles, along with prospects of AI for host–pathogen interactions (HPI), are discussed.
Keywords: virus, deep learning, virus-cell interaction, sequence analysis, computer vision, natural language processing, NLP, DL
Introduction
The causative agents of infectious diseases come in a great variety of shapes, biochemistry, and genetic makeup. They originate from a variety of population reservoirs and only cross their barriers on occasion,1,2 due to the subtle changes in the dynamic ecological equilibrium. In the face of climate changes of as yet unseen proportions, the change in ecological balances will inevitably cause emergence of new infectious diseases.3,4 This makes further pandemics highly plausible. To tackle the outbreaks of the future we must improve our understanding of infectious diseases caused by the interactions between a pathogen and a host. In this review we will explore how novel techniques for computational analysis of interactions between a pathogen and its host may foster such understanding. For the purpose of this review, we will focus primarily on viruses, however some aspects of interactions between viruses and their hosts may be generalized to other pathogens.
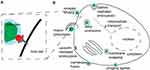
Viruses require host cells to procreate and spread their progeny. For this, they enter cells, replicate and egress in a stepwise process occurring through interactions between molecules of the host cell and the pathogen molecules. Such interactions are commonly referred to as host–pathogen interactions (HPI).5 HPIs include mediating various host mechanisms’ exploitation by the pathogen. Typically, this occurs through direct interactions between the molecules of the pathogen and the molecules of the host cell (Figure 1A). For example, SARS-CoV2 virus enters human host cells through the binding of its S protein to Angiotensin-converting Enzyme 2 (ACE-2) of the cells. Next, in brief, particles get endocytosed, fuse with endosomes and uncoat their genome, perform primary protein translation, and viral RNA synthesis. Consecutively, virus forms replication factories, producing progeny building blocks, and assembles progeny, which subsequently egress the infected host-cell.6 Needless to say, on a mechanistic level this process involves dozens of HPIs which give the virus its advantages and may be exploited as antiviral strategies. Additionally, these mechanisms vary greatly for other viruses7 (Figure 1B) and even other human coronaviruses (HCoV).
For example, S proteins of the predominant seasonal HCoV-OC43 (Betacoronavirus 1) and HCoV-229E (Alphacoronavirus)8,9 bind to 9-O-acetylsialic acids and Amino-peptidase N as primary receptors in human cells rather than ACE-2. Similarly, the primary receptor of the HCoV-HKU1 responsible for the first severe acute respiratory syndrome (SARS) outbreak is 9-O-acetylsialic acids rather than ACE-2.10 In other words, human pathogenic viruses constantly change and adapt. Therefore, understanding biological mechanisms of virus entry, replication, and egress may allow to develop general strategies for fighting infectious diseases. However, identifying HPIs on a mechanistic level is often a laborious manual experimental task. Furthermore, due to the ability of the virus to change and adapt, deciphering HPI mechanisms in a timely fashion is akin to chasing a moving target. In the currently ongoing SARS-CoV2 outbreak, identifying important mechanistic changes in the emerging virus variants fast is the question of life and death. Perhaps the most promising weapon to tackle laborious manual tasks that humanity has in its arsenal to date are machine learning (ML) and artificial intelligence (AI). While experimental validation remains a must, these computational techniques may significantly narrow down the number of experiments required for HPI identification through predicting putative interaction partners. Such HPI prediction tasks include predicting host–pathogen protein–protein interactions11,12 (e.g., using protein sequences and infection phenotypes13), prediction of a putative host or a receptor for the specific pathogen.14
Historically, the field of AI originates from an attempt to create fundamental and applied basics for machines with “intelligent properties”.15 ML, often considered an AI subfield, has significantly facilitated AI by providing a tangible toolset. Conversely, the traction some ML algorithms like artificial neural networks have gained in recent years may be to a larger extent attributed to the effort to make progress in AI. Taken together, these methodologies refer to a group of computational techniques that enable computers to perform specific tasks without the use of explicit rules or instructions.16 This is usually accomplished by creating ML models from real world or computer simulated data. Typically, an ML model is trained on a data set of engineered features (e.g., extraction of cell size, signal intensity, etc.) in a user-supervised or unsupervised manner. Conventionally, a supervised data set consists of the features (X) and targets (Y), where Y corresponds to an objectively known property (i.e., the ground truth). An approach like this is well suited to automating biological data processing, where formulating a detailed and finite set of rules denoting related events is difficult.17–19
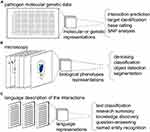
Beyond classical ML, a subset of approaches named Deep Learning (DL) uses algorithms like deep artificial neural networks (DNN) to provide a methodology for pattern recognition with unprecedented accuracy.20 This is achieved through a combination of approaches including representation learning, allowing automated feature generation,21 as well as, stacking multiple hidden layers (modules) of artificial neural networks. Combining various kinds of connections and linear algebra operations between individual neurons in a layer allows to construct a broad variety of DNN layers including, for example, convolutional layers,22 recurrent layers,23 attention layers,24 Sigmoid or SoftMax classification layers. Modern DNN architectures reach expressive capacity of billions or even trillions25 trainable parameters.26 Noteworthy, as a rule of thumb, the larger the expressive capacity of a DNN the more data points are required to train while avoiding overfitting. This bears design choice constraints in the domain specific fields like HPI analysis. To facilitate training of such advanced architectures, Practicality of DL relies on the modern parallel computing, which allows it to represent the features of the input data (e.g., micrographs or CT scans) through non-linear transformations in the so-called vector latent space. Recently, adoption of these methodologies has dramatically increased in the field of Infections Biology. Applications of AI, ML and DL span molecular and genetic data (Figure 2A), microscopy (Figure 2B) and language data (Figure 2C). Here we will review specific examples of these applications and discuss the future outlook for ML and AI for the prediction of HPIs.
Host–Pathogen Interactions Analysis from Genetic and Molecular Data
Perhaps one of the most direct applications of AI and ML for HPIs is to reveal patterns on the level of host and pathogen molecules and genes. The genetic or molecular information is typically represented on a single-character sequence level (Figure 2A). Small molecule information is often represented using a simplified molecular-input line-entry system (smiles).27 In such a setting ML can assist in RNA and DNA accessibility analysis, transcription analysis (reviewed in28), protein–protein interactions,11–13,29 as well as, sequence-based host organism or receptor prediction.14 Noteworthy, in well-defined tasks simple ML algorithms like Random Forest (RF)30 classification, Multilayer Perceptron (MLP) or kernel-based SVM31 perform remarkably well. For example, Karabulut et al show that on the task of adenovirus infection genus prediction kernel-based SVM reaches performance of 0.96 F1 score and 0.89 area under the receiver operating characteristics curve (AUC) with RF and MLP algorithms trailing remarkably close.29 In such cases, more advanced algorithms like DL are not very likely to deliver a significant further improvement. However, in the settings outside of the very specific data set these algorithms may deliver a boost in generalization.
Identification of genetic variations in either host or pathogen genomes conferring higher pathogenicity may also be improved using DL.28 Other examples of successful ML application for HPIs include base calling and SNP analysis32 and clinical metagenomics.33 Algorithms of choice most commonly include recurrent neural networks (RNN), for example architecture known as long short-term memory (LSTM) RNN.23 In some cases, more specialized CNNs are employed34 or even a consecutive combination of CNN and LSTM.35 Traditionally, RNN (and sometimes CNN) architectures have dominated the algorithmic landscape for sequence-based analysis, demonstrating state-of-the-art performance. However, recently introduced transformer architecture,26 which is currently dominating natural language ML, is slowly entering the field.36 Yet data-hungriness remains the biggest hurdle for the transformers to overcome in the HPI domain.
Beyond simple pattern recognition AI and ML algorithms trained on a large number of examples may be used to generate putative new molecules. This approach is showing promising results in the novel antiviral space. Beck and colleagues, for example, described prospective antivirals for the pandemic SARS-CoV2 using a DL model of drug-target interaction from a large number of commercially available antivirals. For this, they developed the Molecule Transformer - Drug Target Interaction architecture. Using this technique, authors identified human immunodeficiency virus drug as a potential candidate.37
Host–Pathogen Interactions Analysis from Image Data
HPIs may be observed visually or using digital microscopy. On a subcellular level microscopic imaging may be employed to capture image-based data of individual virus particle interactions with host-cell proteins.38,39 These data are typically obtained using fluorescence light microscopy at high magnification (e.g., 64x-100x), supperresolution microscopy38,40,41 or electron microscopy.42–44 Various ML techniques may be employed to analyze HPIs subcellular data, ranging from support vector machines45 to DL.46–50
Infection manifestation on a single-cell level is typically hallmarked by the onset of cytopathic effect (CPE).51,52 Synchronized with virus entry, uncoating, and replication through a virus genetic program, virus-induced CPE involves dramatic changes of cell morphology,51,52 which can be observed in cell culture using conventional light microscopy. These include, among other, cell rounding and swelling, focal patterns emergence, cytoplasmic vacuolization53 pyknosis (cytoplasmic shrinkage),51 syncytia formation,54 and may be linked with pathogen-related cell death,51 apoptosis55 or motility.56 The HPIs occurring on molecular level drive CPE. CPE can be observed in cell culture using conventional light microscopy techniques without specific labeling at a moderate magnification (e.g., 5×-20×). Its manifestation often differs substantially for various pathogens, cell types, and multiplicity of infection (MOI).57
Downstream tasks (Figure 2B) for which ML is employed on such data typically include pathogen image segmentation49 (often using a variety of the U-Net architecture58), HPI events or virus classification from full or cropped field-of-view,47,48,50 pathogen object detection59 or infection detection.60 Other examples include understanding structure and function relationships with the pathogens40 or time-lapse analysis.45,60
Being strongly related to the field of computer vision, HPI image analysis remains, thus far, strongly dominated by the CNN DL algorithms. Indeed, unsurprisingly for pathogen image classification tasks both shallower and deeper CNNs outperform conventional RF and MLP ML algorithms in metrics like F1 often by more than 30–40%.47,60
Host–Pathogen Interactions Analysis from the Language Data in the Scientific Publications
ML for natural language processing (NLP) has recently seen an incredible boost in performance through the introduction of the transformer-based models.26 Specifically, in their work Devlin et al reported that BERT transformer mode significantly outperformed bidirectional LSTM (state-of-the-art at that time) on the General Language Understanding Evaluation (GLUE)61 benchmark with 71 and 82 average GLUE performance, Transformer models leverage large text corpora, akin to BookCorpus62 or the English Wikipedia data set, and high expressive capacity to define the new state-of-the-art performance on a plethora of NLP tasks. These tasks include text classification, named entity recognition (NER), semantic text similarity (STS), text summary, question answering (QA), reading comprehension, knowledge discovery (KD) and mapping and other (reviewed in63). Further boost in performance in the novel transformer architectures is achieved through the multi-headed attention mechanism.24 Building upon it through transfer learning (i.e., repurposing a pretrained model), the general-purpose deep bidirectional transformers model was fine-tuned by Lee and colleagues to the domain of biomedical research texts.64 This work, in turn, sparked a surge in biomedical NLP research.
With respect to HPI research, adoption of this novel methodology was limited by the availability of data sets large-enough to warrant a successful domain adaptation until a few years ago. However, amid the global SARS-CoV2 pandemic Wang and colleagues constructed the so-called COVID-19 open research data set (CORD-19).65 This data set, in turn, inspired a plethora of HPI analysis approaches using the language data in scientific publications. A great number of NLP downstream tasks have been attempted on this and similar data sets in the past 12–18 month alone (Figure 2C).
Specifically, Koksal et al proposed a search engine approach to finding protein-compound pairs in COVID-19 literature formulated in the CORD-19 corpus.66 Wang and co-authors have formulated a NER task data set that covers 75 detailed entity types.67 These types include biomedical entities like genes, chemicals and diseases, as well as, entity types related explicitly to the SARS-CoV2 and COVID-19 research including coronaviruses, viral proteins, materials, evolution, immune responses and substrates. To capture subtle domain specific similarities in the CORD-19 data set Guo et al formulated an STS data set.68 For the KD task on CORD-19 data set Tam and co-authors proposed a transformer-based target-query method they named Transformer Query-Target Knowledge Discovery (TEND).69 Reddy and colleagues proposed a QA task domain adaptation for COVID-19 related questions70 using a combination of CORD-19 and several COVID-19 QA data sets.71–73 Their data suggest that pretrained models may successfully be fine-tuned for HPI domain, gaining 4–7% performance over the baseline. Noteworthy, many of these recent approaches remain to be peer-reviewed and the impact these techniques will have on the SARS-CoV2 and HPI research remains to be demonstrated.
Conclusion
Prior to the surge of AI-community interest in the biomedical domain, ML techniques have been only sporadically applied in analysis of interactions between pathogens and their hosts (reviewed in74). However, in the wake of SARS-CoV2 pandemic ML and AI techniques application for HPI analysis sees an ever-growing interest. Transcending the status of a niche application of novel AI algorithms, pathogens and HPI research found itself in the spotlight of AI-researchers attention. This manifested in a variety of fundamentally new approaches to analyze molecular, image-based and more recently language-based HPI data mentioned in this review. Admittedly, the knowledge gap between AI and HPI research remains great. This is especially evident in the case of language-based data. Yet, given the sheer effort from both fields to bridge this gap, we may see fruition of these efforts in a not-so-distant future.
The lack of large HPI data sets remains one of the main hurdles for further penetration of AI and ML into the HPI field. However, recent years have seen a significant improvement in that matter. In case of image-based data, the advent of resources like Bio-Image Archive,75 data-dedicated journals and social coding platforms fostered deposition of specialized image data sets, including those focused on HPI.46,76 The influence of CORD-1965 on the adoption of NLP techniques in HPI research was unequivocally self-evident. Perhaps, it is the access to open research data sets akin to CORD-19 together with image-based data repositories that will become the missing piece for AI and ML to become a viral case in HPI research.
Abbreviations
ACE-2, angiotensin-converting enzyme 2; AI, artificial intelligence; AUC, area under receiver operating characteristics curve; COVID-19, coronavirus disease 2019; CORD-19, COVID-19 open research data set; CT, computed tomography; DL, deep learning; DNN, deep artificial neural networks; EM, electron microscopy; GLUE, General Language Understanding Evaluation; HCoV, human coronaviruses; HPI, host–pathogen interactions; KD, knowledge discovery; ML, machine learning; NLP, natural language processing; NER, named entity recognition; SARS-CoV2, severe acute respiratory syndrome coronavirus 2; STS, semantic text similarity; TEND, transformer query-target knowledge discovery; QA, question answering.
Disclosure
The author reports no conflicts of interest in this work.
References
1. Shortridge K. Pandemic influenza: a zoonosis? Semin Respir Infect. 1992;7:11–25.
2. Hahn BH, Shaw GM, De KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287(5453):607–614. doi:10.1126/science.287.5453.607
3. Patz JA, Graczyk TK, Geller N, Vittor AY. Effects of environmental change on emerging parasitic diseases. Int J Parasitol. 2000;30(12–13):1395–1405. doi:10.1016/S0020-7519(00)00141-7
4. Patz JA, Reisen WK. Immunology, climate change and vector-borne diseases. Trends Immunol. 2001;22(4):171–172. doi:10.1016/S1471-4906(01)01867-1
5. Casadevall A, Pirofski LA. Host-pathogen interactions: basic concepts of microbial commensalism, colonization, infection, and disease. Infect Immun. 2000;68(12):6511–6518. doi:10.1128/IAI.68.12.6511-6518.2000
6. V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2020;19:155–170.
7. Yamauchi Y, Helenius A. Virus entry at a glance. J Cell Sci. 2013;126(6):1289–1295.
8. Lau SK, Lee P, Tsang AK, et al. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J Virol. 2011;85(21):11325–11337. doi:10.1128/JVI.05512-11
9. Gaunt ER, Hardie A, Claas EC, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48(8):2940–2947. doi:10.1128/JCM.00636-10
10. Guruprasad L. Human coronavirus spike protein-host receptor recognition. Prog Biophys Mol Biol. 2020.
11. Zheng N, Wang K, Zhan W, Deng L. Targeting virus-host protein interactions: feature extraction and machine learning approaches. Curr Drug Metab. 2019;20(3):177–184. doi:10.2174/1389200219666180829121038
12. Cuesta-Astroz Y, Oliveira G. Computational and experimental approaches to predict host–parasite protein–protein interactions. In: Computational Cell Biology. New York, NY: Humana Press; 2018:153–173.
13. Liu-Wei W, Kafkas S, Chen J, Dimonaco NJ, Tegnér J, Hoehndorf R. DeepViral: prediction of novel virus-host interactions from protein sequences and infectious disease phenotypes. 2021.
14. Mock F, Viehweger A, Barth E, Marz M. VIDHOP, viral host prediction with Deep Learning. Bioinformatics. 2021;37(3):318–325. doi:10.1093/bioinformatics/btaa705
15. Fisch DH, Yakimovich A, Clough B, et al. An Artificial Intelligence Workflow for Defining Host-Pathogen Interactions. bioRxiv. 2018:408450.
16. Mitchell TM. Machine learning. 1997.
17. Tarca AL, Carey VJ, Chen XW, Romero R, Drăghici S. Machine learning and its applications to biology. PLoS Comput Biol. 2007;3(6):e116. doi:10.1371/journal.pcbi.0030116
18. Sommer C, Gerlich DW. Machine learning in cell biology–teaching computers to recognize phenotypes. J Cell Sci. 2013;126(24):5529–5539.
19. Sommer C, Straehle C, Kothe U, Hamprecht FA. ilastik: interactive learning and segmentation toolkit.
20. LeCun Y, Bengio Y, Hinton G. Deep learning. nature. 2015;521(7553):436. doi:10.1038/nature14539
21. Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell. 2013;35(8):1798–1828. doi:10.1109/TPAMI.2013.50
22. LeCun Y, Bengio Y. Convolutional networks for images, speech, and time series. In: Arbib MA, editor. The Handbook of Brain Theory and Neural Networks. MIT Press; 1995:3361.
23. Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi:10.1162/neco.1997.9.8.1735
24. Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. Adv Neural Inf Process Syst. 2017;30:5998–6008.
25. Fedus W, Zoph B, Shazeer N. Switch transformers: scaling to trillion parameter models with simple and efficient sparsity. arXiv Preprint arXiv:210103961. 2021.
26. Devlin J, Chang MW, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv Preprint arXiv:181004805. 2018.
27. Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci. 1988;28(1):31–36.
28. Zou J, Huss M, Abid A, Mohammadi P, Torkamani A, Telenti A. A primer on deep learning in genomics. Nat Genet. 2019;51(1):12–18. doi:10.1038/s41588-018-0295-5
29. Karabulut OC, Karpuzcu BA, Türk E, Ibrahim AH, Süzek BE. ML-AdVInfect: a machine-learning based adenoviral infection predictor. Front Mol Biosci. 2021;8. doi:10.3389/fmolb.2021.647424
30. Ho TK. Random decision forests.
31. Crammer K, Singer Y. On the algorithmic implementation of multiclass kernel-based vector machines. J Mach Learn Res. 2001;2:265–292.
32. Poplin R, Chang P-C, Alexander D, et al. A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol. 2018;36(10):983–987. doi:10.1038/nbt.4235
33. Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341. doi:10.1038/s41576-019-0113-7
34. Tampuu A, Bzhalava Z, Dillner J, Vicente VR, Melcher U. ViraMiner: deep learning on raw DNA sequences for identifying viral genomes in human samples. PLoS One. 2019;14(9):e0222271. doi:10.1371/journal.pone.0222271
35. Veltri D, Kamath U, Shehu A. Deep learning improves antimicrobial peptide recognition. Bioinformatics. 2018;34(16):2740–2747. doi:10.1093/bioinformatics/bty179
36. Zhang Y, Lin J, Zhao L, Zeng X, Liu X. A novel antibacterial peptide recognition algorithm based on BERT. Brief Bioinform. 2021.
37. Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784–790. doi:10.1016/j.csbj.2020.03.025
38. Gray RD, Albrecht D, Beerli C, et al. Nanoscale polarization of the entry fusion complex of vaccinia virus drives efficient fusion. Nat Microbiol. 2019;4(10):1636–1644. doi:10.1038/s41564-019-0488-4
39. Wang I, Burckhardt CJ, Yakimovich A, Greber UF. Imaging, tracking and computational analyses of virus entry and egress with the cytoskeleton. Viruses. 2018;10(4):166. doi:10.3390/v10040166
40. Gray RD, Beerli C, Pereira PM, et al. VirusMapper: open-source nanoscale mapping of viral architecture through super-resolution microscopy. Sci Rep. 2016;6:29132. doi:10.1038/srep29132
41. Yakimovich A, Huttunen M, Samolej J, et al. Mimicry embedding for advanced neural network training of 3D biomedical micrographs. bioRxiv. 2019:820076.
42. Dales S, Siminovitch L. The development of vaccinia virus in Earle’s L strain cells as examined by electron microscopy. J Biophys Biochem Cytol. 1961;10(4):475–503. doi:10.1083/jcb.10.4.475
43. Dales S, Eggers HJ, Tamm I, Palade GE. Electron microscopic study of the formation of poliovirus. Virology. 1965;26:379–389. doi:10.1016/0042-6822(65)90001-2
44. Nii S, Morgan C, Rose HM. Electron microscopy of herpes simplex virus: II. Sequence of development. J Virol. 1968;2(5):517–536. doi:10.1128/jvi.2.5.517-536.1968
45. Wang I-H, Burckhardt CJ, Yakimovich A, Morf MK, Greber UF. The nuclear export factor CRM1 controls juxta-nuclear microtubule-dependent virus transport. J Cell Sci. 2017;130(13):2185–2195.
46. Georgi F, Kuttler F, Murer L, et al. A high-content image-based drug screen of clinical compounds against cell transmission of adenovirus. Scientific Data. 2020;7(1):1–12. doi:10.1038/s41597-020-00604-0
47. Fisch D, Yakimovich A, Clough B, et al. Defining host–pathogen interactions employing an artificial intelligence workflow. eLife. 2019;8:e40560. doi:10.7554/eLife.40560
48. Nanni L, De Luca E, Facin ML, Maguolo G. Deep learning and handcrafted features for virus image classification. J Imag. 2020;6(12):143. doi:10.3390/jimaging6120143
49. Matuszewski DJ, Sintorn I-M. Reducing the u-net size for practical scenarios: virus recognition in electron microscopy images. Comput Methods Programs Biomed. 2019;178:31–39. doi:10.1016/j.cmpb.2019.05.026
50. Zhang L, Yan WQ. Deep learning methods for virus identification from digital images.
51. Agol VI. Cytopathic effects: virus-modulated manifestations of innate immunity? Trends Microbiol. 2012;20(12):570–576. doi:10.1016/j.tim.2012.09.003
52. Mocarski ES, Upton JW, Kaiser WJ. Viral infection and the evolution of caspase 8-regulated apoptotic and necrotic death pathways. Nat Rev Immunol. 2012;12(2):79. doi:10.1038/nri3131
53. Shubin AV, Demidyuk IV, Komissarov AA, Rafieva LM, Kostrov SV. Cytoplasmic vacuolization in cell death and survival. Oncotarget. 2016;7(34):55863. doi:10.18632/oncotarget.10150
54. Suchman E, Blair C Cytopathic effects of viruses protocols. 2007.
55. Shen Y, Shenk TE. Viruses and apoptosis. Curr Opin Genet Dev. 1995;5(1):105–111. doi:10.1016/S0959-437X(95)90061-6
56. Beerli C, Yakimovich A, Kilcher S, et al. Vaccinia virus hijacks EGFR signalling to enhance virus spread through rapid and directed infected cell motility. Nat Microbiol. 2018;4:216–225.
57. González-Sánchez H, Monsiváis-Urenda A, Salazar-Aldrete C, et al. Effects of cytomegalovirus infection in human neural precursor cells depend on their differentiation state. J Neurovirol. 2015;21(4):346–357. doi:10.1007/s13365-015-0315-5
58. Ronneberger O, Fischer P, Brox T U-net: convolutional networks for biomedical image segmentation.
59. Ito E, Sato T, Sano D, Utagawa E, Kato T. Virus particle detection by convolutional neural network in transmission electron microscopy images. Food Environ Virol. 2018;10(2):201–208. doi:10.1007/s12560-018-9335-7
60. Andriasyan V, Yakimovich A, Petkidis A, et al. Microscopy deep learning predicts virus infections and reveals mechanics of lytic-infected cells. Iscience. 2021;24(6):102543. doi:10.1016/j.isci.2021.102543
61. Wang W, Yan M, Wu C. Multi-granularity hierarchical attention fusion networks for reading comprehension and question answering. arXiv Preprint arXiv:181111934. 2018.
62. Zhu Y, Kiros R, Zemel R, et al. Aligning books and movies: towards story-like visual explanations by watching movies and reading books.
63. Gillioz A, Casas J, Mugellini E, Abou Khaled O. Overview of the Transformer-based Models for NLP Tasks.
64. Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234–1240.
65. Wang LL, Lo K, Chandrasekhar Y, et al. CORD-19: the covid-19 open research dataset. ArXiv. 2020.
66. Köksal A, Dönmez H, Özçelik R, Ozkirimli E, Özgür A. Vapur: a search engine to find related protein–compound pairs in COVID-19 literature. arXiv Preprint arXiv:200902526. 2020.
67. Wang X, Song X, Li B, Guan Y, Han J. Comprehensive named entity recognition on cord-19 with distant or weak supervision. arXiv Preprint arXiv:200312218. 2020.
68. Guo X, Mirzaalian H, Sabir E, Jaiswal A, Abd-Almageed W. Cord19sts: covid-19 semantic textual similarity dataset. arXiv Preprint arXiv:200702461. 2020.
69. Tam LK, Wang X, Xu D. Transformer query-target knowledge discovery (TEND): drug discovery from CORD-19. arXiv Preprint arXiv:201204682. 2020.
70. Reddy RG, Iyer B, Sultan MA, et al. End-to-end QA on COVID-19: domain adaptation with synthetic training. arXiv Preprint arXiv:201201414. 2020.
71. Möller T, Reina A, Jayakumar R, Pietsch M. COVID-QA: a question answering dataset for COVID-19.
72. Tang R, Nogueira R, Zhang E, et al. Rapidly bootstrapping a question answering dataset for COVID-19. arXiv Preprint arXiv:200411339. 2020.
73. Lee J, Yi SS, Jeong M, et al. Answering questions on covid-19 in real-time. arXiv Preprint arXiv:200615830. 2020.
74. Sen R, Nayak L, De RK. A review on host–pathogen interactions: classification and prediction. Eur J Clin Microbiol Infect Dis. 2016;35(10):1581–1599. doi:10.1007/s10096-016-2716-7
75. Ellenberg J, Swedlow JR, Barlow M, et al. A call for public archives for biological image data. Nat Methods. 2018;15(11):849–854. doi:10.1038/s41592-018-0195-8
76. Yakimovich A, Huttunen M, Samolej J, et al. Mimicry embedding facilitates advanced neural network training for image-based pathogen detection. Msphere. 2020;5(5):e00836–20. doi:10.1128/mSphere.00836-20
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.