Back to Journals » International Journal of Nanomedicine » Volume 18
M2 Macrophage-Derived Exosomal lncRNA MIR4435-2HG Promotes Progression of Infantile Hemangiomas by Targeting HNRNPA1
Authors Li Z , Cao Z, Li N , Wang L, Fu C, Huo R , Xu G , Tian C, Bi J
Received 25 August 2023
Accepted for publication 7 October 2023
Published 20 October 2023 Volume 2023:18 Pages 5943—5960
DOI https://doi.org/10.2147/IJN.S435132
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lijie Grace Zhang
Zhiyu Li,1 Zhongying Cao,1 Nanxi Li,2 Luying Wang,2 Cong Fu,2 Ran Huo,1,2 Guangqi Xu,2 Chonglin Tian,2,* Jianhai Bi1– 3,*
1Department of Plastic and Aesthetic Surgery, Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University, Jinan, People’s Republic of China; 2Department of Plastic and Aesthetic Surgery, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, People’s Republic of China; 3Medical Science and Technology Innovation Center, Shandong First Medical University & Shandong Academy of Medical Sciences, Jinan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianhai Bi, Department of Plastic and Aesthetic Surgery, Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University, No. 324, Jingwu Road, Huaiyin District, Jinan, Shandong Province, 250021, People’s Republic of China, Tel +86-15110080702, Email [email protected] Chonglin Tian, Department of Plastic and Aesthetic Surgery, Shandong Provincial Hospital Affiliated to Shandong First Medical University, No. 324, Jingwu Road, Huaiyin District, Jinan, Shandong Province, 250021, People’s Republic of China, Tel +86-15666966900, Email [email protected]
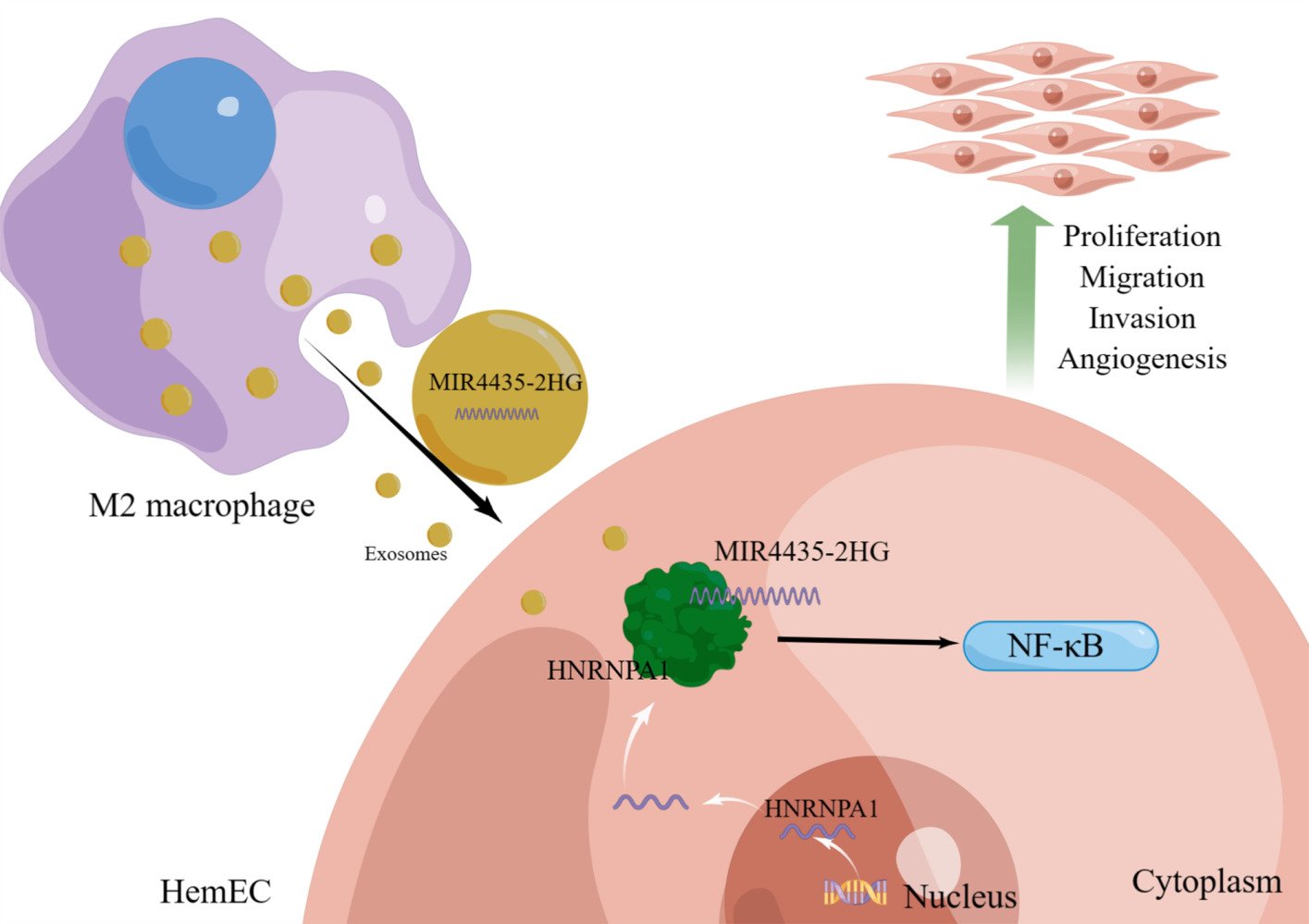
Purpose: Infantile hemangiomas (IHs) are commonly observed benign tumors that can cause serious complications. M2-polarized macrophages in IHs promote disease progression. In this study, we investigated the role of M2 macrophage-derived exosomal lncRNA MIR4435-2HG in IHs.
Patients and Methods: Exosomes derived from M2 polarized macrophages were extracted. Next, using cell co-culture or transfection, we investigated whether M2 polarized macrophage-derived exosomes (M2-exos) can transport MIR4435-2HG to regulate the proliferation, migration, invasion, and angiogenesis of hemangioma-derived endothelial cells (HemECs). RNA-seq and RNA pull-down assays were performed to identify targets and regulatory pathways of MIR4435-2HG. We explored the possible mechanisms through which MIR4435-2HG regulates the biological function of HemECs.
Results: M2-exos significantly enhanced the proliferation, migration, invasion, and angiogenesis of HemECs. Thus, HemECs uptake M2-exos and promote biological functions through the inclusion of MIR4435-2HG. RNA-seq and RNA pull-down experiments confirmed that MIR4435-2HG regulates of HNRNPA1 expression and directly binds to HNRNPA1, consequently affecting the NF-κB signal pathway.
Conclusion: MIR4435-2HG of M2-exos promotes the progression of IHs and enhances the proliferation, migration, invasion, and angiogenesis of HemECs by directly binding to HNRNPA1. This study not only reveals the mechanism of interaction between M2 macrophages and HemECs, but also provides a promising therapeutic target for IHs.
Keywords: hemangioma-derived endothelial cell, infantile hemangioma, M2-polarized macrophage, MIR4435-2HG, HNRNPA1
Graphical Abstract:
Introduction
Infantile hemangiomas (IHs) are the most commonly observed soft tissue tumors of the skin during infancy, with an incidence rate of approximately 4.5%.1 IHs appear shortly after birth and exhibit a unique life cycle characterized by rapid growth during the first 9–12 months, followed by slow and spontaneous regression.2,3 Complications, including obstruction, dysfunction, ulceration, and disfigurement, occur in approximately 10% of patients.4 Site-specific IHs can cause significant problems, such as organ failure, visual impairment, limited joint movement, respiratory distress, and death.5 Although oral propranolol is clinically effective in the treatment of IHs, the mechanisms of proliferation and regression in IHs remain unclear.6 In addition, resistance to propranolol and the possibility of treatment failure still exist, especially in patients with high risk IHs.7 Therefore, studying the pathogenesis of IHs to develop targeted therapies is critical.
Macrophages are heterogeneous cells present in all organ systems and play notable roles in innate and adaptive immunity, hematopoiesis, angiogenesis, reproduction, and systemic metabolism.8 Macrophages respond differently to different stimuli in vivo and in vitro and exhibit significant functional differences. Macrophages are usually divided into two subtypes: M1 (classically activated macrophages) and M2 (alternatively activated macrophages).9 M2-polarized macrophages promote the proliferation and endothelial differentiation of hemangioma-derived stem cells (HemSCs) and inhibit their adipogenesis.10,11 Exosomes are vesicles 30–150 nm in diameter produced by various cells and carry various bioactive substances (eg, proteins, lipids, DNA, and RNA).12 Exosomes are closely associated with IHs. MiRNA-196b-5p, derived from exosomes in HemSCs, promotes the proliferation and angiogenesis of hemangioma-derived endothelial cells (HemECs) and attenuates the apoptosis and cell cycle inhibition rate of HemECs.13 M2 polarized macrophage-derived exosomes (M2-exos) deliver miR-27a-3p from macrophages to HemSCs to reduce the sensitivity of HemSCs to propranolol.14 However, M2-exos and IHs have only been explored up to the miRNA level. Therefore, refinement of the related mechanisms requires further exploration.
LncRNAs are RNA molecules that are 200 nucleotides or longer and do not encode proteins.15 LncRNAs are involved in several physiological processes because they regulate gene expression at the epigenetic, transcriptional, and post-transcriptional levels.16 Aberrant expression is also closely associated with tumor proliferation, differentiation, apoptosis, and metastasis.17 The lncRNA MIR4435-2 host gene (MIR4435-2HG) is located on human chromosome 2q13. Its expression is upregulated in 18 tumors and is involved in six signaling pathways that promote tumorigenesis.18 MIR4435-2HG promotes the progression of various tumors, such as those of liver, breast, and bladder cancers,19–21 and MIR44435-2HG is indirectly involved in numerous biological functions through the molecular mechanism of competitive endogenous RNA network (ceRNA).22 Compared with the indirect mechanism of ceRNAs, it is more anticipated that MIR44435-2HG regulate proteins by directly binding to them; however, its expression and mechanism in IHs remain unclear.
In this study, we demonstrated that M2-exos effectively promoted the proliferation, migration, invasion, and angiogenesis of HemECs. M2-exos promoted the growth of IHs in vitro and in vivo by transporting MIR44435-2HG. We found that MIR44435-2HG directly bind to heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1) in HemECs to regulate cellular biological functions. Therefore, we explored the potential role of MIR44435-2HG in IHs and attempted to reveal the link between M2-exos and IH progression.
Materials and Methods
Ethical Statement
The human subject protocol was approved by the Biomedical Research Ethics Committee of Shandong Provincial Hospital (SZRJJ:NO.2021–414). All clinical samples were obtained from previously untreated patients with IH at Shandong Provincial Hospital (Shandong, China). Informed consent was obtained from the parents/guardians of all participants in accordance with the World Medical Association Declaration of Helsinki. All animal studies were performed in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals and were approved by the Institutional Laboratory Animal Ethics Committee of Shandong Provincial Hospital (NO.2022–061).
Induction and Identification of Macrophage Polarization
THP-1 cells and the culture medium were purchased from Procell (Wuhan, China; 1×106cells, third generation; Identified through STR; No Mycoplasma contamination detected). THP-1 cells were inoculated in medium and cultured at 37 °C, 5% CO2, and 90% humidity in an incubator. THP-1 cells were seeded in 6-well plates, and M0 macrophages were stimulated with 100 ng/mL phorbol 12-myristate 13-acetate (PMA, Solarbio, China) for 48 h. M0 macrophages were incubated with 20 ng/mL IL-4 and 20 ng/mL IL-13 (Solarbio) for 48 h to induce M2 polarization. Macrophages were collected, and subsequently subjected to qRT-PCR analysis to detect CD11b, CD14, CD68, iNOS, IL-10, and CD206 to identify M0 and M2 macrophages (Table 1 for primer sequences).
 |
Table 1 Primers Used in This Manuscript |
Extraction of M0 and M2 Macrophage Exosomes
M0 and M2 macrophages were cultured in exosome-free serum for 48 h. The collected supernatant was centrifuged at 500 g for 10 min to remove cellular contamination. The supernatant was further centrifuged at 12,000 g for 20 min to remove apoptotic bodies and large cellular debris. The supernatant was centrifuged at 100,000 g for 90 min to enrich the exosomes and then rinsed with phosphate buffered saline (PBS). Finally, exosomes were collected via ultracentrifugation at 100,000 g for 90 min and resuspended in an appropriate amount of PBS.
Identification and Uptake of Exosomes
The exosomes (10 μL) were collected dropwise and precipitated on a copper grid for 1 min, and the floating solution was blotted off using a filter paper. Phosphotungstic acid (10 μL) was added dropwise on the copper net and precipitated for 1 min, and the floating solution was removed through aspiration on filter paper. After drying at room temperature for a few minutes, 100 kv was imaged for transmission electron microscopy (TEM). Nanoparticle tracking analysis was performed to determine the size and concentration of the exosomes. The expression of exosome markers, HSP70, TSG101, and CD9 (calnexin is a cytoplasmic endoplasmic reticulum-specific protein), was analyzed via the Western blotting of M0-exos and M2-exos and the corresponding supernatants. M2-exos was added to HemECs medium at 100 μg/mL, and EvLINK and CellLINK double staining (Tianjiu, Tianjin, China) were used to label the exosomes and cell membranes, respectively, to observe the intracellular staining after cellular phagocytosis of exosomes.
Cell Extraction and Culture
HemECs were extracted from proliferating IHs tissues, as described previously.23 HemECs were cultured in an endothelial cell medium (ScienCell, Shanghai, China), containing 5% fetal bovine serum, 1% penicillin-streptomycin, and 1% endothelial cell growth factor, at 37 °C and 5% CO2.
qRT-PCR
Total RNA was extracted using RNAiso Plus reagent (Takara, Tokyo, Japan). cDNA was reverse transcribed from 1 μg total RNA using HiScript RT Super Mix qPCR (Vazyme, Nanjing, China). The cDNA was quantified using SYBR Green Master Mix (Vazyme) on a Light Cycler 480 II (Roche, Switzerland) for quantitative PCR. The RNA levels were assessed using the 2−ΔΔCT method with β-actin as an internal standard. The primers used in this study are listed in Table 1.
Western Blotting
Total protein was extracted using RIPA lysis buffer (Beyotime, Shanghai, China). Protein concentration was determined using the BCA protein assay kit (Beyotime, Shanghai, China). After separation in 10% SDS-PAGE, proteins were transferred to PVDF membranes (Millipore, Billerica, USA) and incubated with primary antibodies overnight at 4 °C. The antibodies used were as follows: calnexin (90 kDa; rabbit; 1:1000; GB111369; Servicebio, Wuhan, China), HSP70 (70 kDa; rabbit; 1:1000; GB11241; Servicebio), TSG101 (46 kDa; rabbit; 1:1000. GB11618; Servicebio), CD9 (25 kDa; Rabbit; 1:2000; 20597-1-AP; Proteintech), α-tubulin (55 kDa; Rabbit; 1:3000; GB15201; Servicebio), HNRNPA1 (34 kDa; Rabbit; 1:10000; 11176-1-AP; Proteintech), NF-κB p65 (65 kDa; Mouse; 1:2000; 66535-1-lg; Proteintech), phospho-NF-κB p65 (65 kDa; Rabbit; 1:500; GB113882; Servicebio), IkBα (36 kDa; Mouse; 1:10000; 66418-1-lg; Proteintech), and IKB alpha (phospho S36) (35 kDa; Rabbit; 1:10000; ab133462; Abcam). The membranes were then incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:10000; D110058-0100; Sangon Biotech) or Goat Anti-Mouse IgG (H+L) (peroxidase/HRP conjugated)(1:2000; E-AB-1008; Elabscience) for 1 h. A ChemiDoc Imaging System (Bio-Rad, Hercules, CA, USA) was used to quantify protein expression.
Fluorescence in situ Hybridization (FISH)
FISH experiments were performed to detect lncRNA MIR4435-2HG in IHs, normal tissues, and HemECs. A Cy3-labeled MIR4435-2HG probe was designed and synthesized by Servicebio. The 18S probe (Servicebio) was used as a control for cytoplasmic localization, and the U6 probe (Servicebio) was used as a control for nuclear localization. MIR4435-2HG was hybridized overnight with the cells or tissue samples to be tested, according to the manufacturer’s instructions. MIR4435-2HG subcellular localization in the samples was observed using confocal microscopy.
Hematoxylin and Eosin Staining
IHs specimens were formalin-fixed and paraffin-embedded. Dewaxed sections (4 mm) were stained with hematoxylin and eosin.
Immunohistochemical (IHC) Staining
Dewaxed fixed sections were stained with a primary antibody against HNRNPA1 (1:100; 11176-1-AP; Proteintech) and subsequently with a secondary antibody (Servicebio, G1213). The samples were stained using a DAB substrate kit (Servicebio) for 1 min, followed by hematoxylin (Servicebio) for 20s. We scored HNRNPA1 expression in tissues using IHC staining. The scoring criteria for IHC staining were as follows: the extent and intensity of staining in five fields of view were assessed using a microscope (Olympus, Tokyo, Japan) at ×400 magnification. Staining intensity was classified into four grades: no staining, score 0; pale yellow, score 1; pale brown, score 2; and dark brown, score 3. Positive expression areas were classified into five categories: <5%, score 0; 6%–25%, score 1; 26%–50%, score 2; 51%–75%, score 3; and 76%–100%, score 4. The intensity and area scores were multiplied and used as the final HNRNPA1 expression scores. All sections were scored by two independent pathologists at Shandong Provincial Hospital. Patients were blinded to their clinical data. If the scores of the two pathologists differed, the mean score was used for analysis.
Cell Transfection
MIR4435-2HG knockdown (shMIR4435-2HG) and overexpression (OE-MIR4435-2HG), and HNRNPA1 knockdown (shHNRNPA1) and overexpression (OE-HNRNPA1) vectors of the recombinant lentivirus and the corresponding negative control shNC (sh-negative control) and EV (Empty Vector), respectively, were designed and synthesized by GeneChem (Shanghai, China). ShMIR4435-2HG targets the GGTCATTAGGGACAGGCAAAT sequence and ShHNRNPA1 targets the TACCTAGTAGCATAGAGATTT sequence according to 5ʹ–3ʹ direction. GV493 (hU6-MCS-CBh-gcGFP-IRES-puromycin) and GV502 ((polyA-MCS-UBI)RV-SV40-EGFP-IRES-puromycin) were used to construct the knockdown and overexpression lentiviruses of MIR4435-2HG, respectively. shHNRNPA1 targeted AAGCTAGGAGGGAGTGAAATA. GV493 (hU6- MCS-CBh-gcGFP-IRES-puromycin) and CV572 (Ubi-MCS-SV40-Cherry-IRES-neomycin) were used to construct the knockdown and overexpression lentiviruses of HNRNPA1, respectively. Transfection efficiency was assessed based on the expression of green fluorescent protein 72 h after transfection, and the cells were screened with puromycin or neomycin, depending on the vector. Transfection efficiency was tested using qRT-PCR after the first application of each lentivirus.
Cell Counting Kit-8 (CCK-8) Proliferation Assay
Cells were seeded into 96-well plates (Corning, New York, NY, USA). Two hours after inoculation, 10 μL of CCK8 (Elabscience, Wuhan, China) was added to each well. The OD value of each well at 450 nm was measured at 2 h, 4 h, 12 h, 1 d, 2 d, and 3 d.
Wound Healing Assay
HemECs were inoculated into 6-well plates. After 24 h, the wells were gently scraped in the middle using a 200 μL pipette tip. After washing the detached cells with PBS, the HemECs were cultured in a pure medium without serum. Images of the wound at the same location were captured at 0 and 12 h to calculate the migration rate.
Transwell Invasion Assay
A Transwell chamber with an 8-μm pore polycarbonate membrane (Biofil, Guangzhou, China) was used for the invasion assay. A medium containing 10% fetal bovine serum was added to the lower chamber. The cells were suspended in a serum-free medium and inoculated into the upper chamber coated with a matrix gel (ABW, Shanghai, China). After incubation for 24 h, the cells in the upper chamber were scraped with a cotton swab, fixed on the lower surface of the membrane with paraformaldehyde for 30 min, and stained with crystal violet (Solarbio) for 30 min. The cells were counted after capturing the images.
Tube Formation Assay
The 96-well plates coated with matrix gel (ABW) were incubated at 37 °C in 5% CO2. After 30 min, 50 μL of cell suspension was added to each well. Images were captured after 4 h.
RNA Pull-Down
RNA sequences were linearized using the FastDigest XhoI kit (ThermoFisher Scientific, Waltham, MA, USA) and T7 MEGAscript kit (Invitrogen, New York, USA). The sequences were transcribed and purified using the purification kit (TianGen Biotech, Beijing. China). RNA-binding proteins were obtained using a Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific) and identified via mass spectrometry (Shandong Xiuyue Biotechnology Co., Ltd, Jinan, China). All protocols were performed according to the manufacturer’s instructions.
RNA Immunoprecipitation (RIP)
The RIP assay was performed using an RNA immunoprecipitation kit (Millipore), according to the manufacturer’s instructions. Magnetic beads combined with anti-IgG or anti-HNRNPA1 antibodies (ab5832; Abcam, Cambridge, UK) and incubated with total RNA lysates from HemECs. The complexes were washed from the magnetic beads, and the isolated RNA was extracted. MIR4435-2HG expression was detected using qRT-PCR.
RNA Sequence Data Analysis
The Agilent 2100 RNA Nano 6000 assay kit (Agilent Technologies) was used to determine the integrity and concentration of extracted total RNA. Libraries were constructed using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA), according to the manufacturer’s instructions. The transcriptomes of MIR4435-2HG shNC HemECs and shMIR4435-2HG HemECs were analyzed using the Illumina NovaSeq 6000 platform (Shandong Xiuyue Biotechnology Co., Ltd), and two different samples from each group of cells were sequenced.
In vivo Xenograft Experiments
Male BALB/c nude mice (6 weeks old; n = 5) were purchased from GemPharmatech (Jiangsu, China) and maintained in a specific pathogen-free environment. The cells were resuspended in serum-free medium at a concentration of 108 cells/mL and mixed 1:1 with matrix gel (ABW). We injected 200 μL mixture subcutaneously in immunodeficient NOD/SCID mice and removed the tumors after 2 weeks of treatment. The tumor volumes and weights were determined.
Statistical Analysis
All experiments were performed in three independent repetitions. Data were analyzed using the SPSS 25.0 and GraphPad Prism 8 software. Data are expressed as mean ± SD or mean ± SEM. Student’s t-test was used to determine statistical differences between two groups. P < 0.05 was considered statistically significant.
Results
Extraction of Exosomes Secreted by M2-Polarized Macrophages
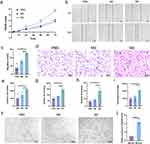
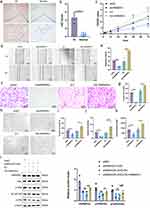
After the induction of THP-1 by PMA, the cell morphology appeared polygonal with a subcircular shape and transformed into anchorage-dependent M0 macrophages (Figure 1a). The morphology of M2 polarized macrophages was similar to that of M0 macrophages and the pseudopods shortened after induction with IL-4 and IL-13. The qRT-PCR results showed that the expression of CD11b, CD14, and CD68 was significantly upregulated in PMA-induced THP-1 cells (Figure 1b). By contrast, no significant difference was observed in iNOS expression between M2 and M0 macrophages, indicating that they were not M1 polarized macrophages, and IL-10 and CD206 expression was significantly upregulated (Figure 1c). These results indicate that we successfully induced M0 and M2 polarized macrophages.
In addition, we isolated M0-exos and M2-exos. TEM showed that vesicles isolated from M2-polarized macrophages were saucer-shaped (Figure 1d), with an average particle size of approximately 77.48 nm (Figure 1e). In addition, Western blotting results showed that M0- and M2-exos expressed the exosome markers CD9, TSG101, and HSP70, whereas CD9, TSG101, and HSP70 were not expressed or were less expressed in the cell lysate (Figure 1f). Calnexin was expressed in the cell lysate, but not in M0- and M2-exos. These results indicate that we successfully isolated M0- and M2-exos. HemECs were co-cultured with M2-exos for 24 h, and the uptake of M2-exos by HemECs was observed using EvLINK and CellLINK double staining assays (Figure 1g).
Functional Regulation of HemECs by M2-Exos and lncRNA MIR4435-2HG Expression
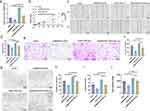
To investigate whether M2-exos can regulate HemEC function, we added PBS, M0-exos, and M2-exos to the HemEC medium. The CCK-8 assay showed that HemEC proliferation was enhanced more in the M2-exos group than that in the PBS and M0-exos groups (Figure 2a). Cell migration ability was confirmed using wound healing assay, which showed enhanced migration of HemECs in the M2-exos group (Figure 2b and c). The Transwell invasion assay showed an enhanced invasion ability of HemECs in the M2-exos group (Figure 2d and e). The tube formation assay showed enhanced tube formation ability of HemECs in the M2-exos group, with an increase in the number of junctions and meshes and in total segments length (Figure 2f–i). The transcript levels of MIR4435-2HG were determined using qRT-PCR. The results showed that MIR4435-2HG was expressed in the HemECs of the M2-exos group compared to that in the M0-exos group (Figure 2j).
MIR4435-2HG Expression Increases in IHs and Regulates Biological Function of IH in vivo and in vitro
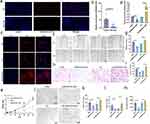
The clinical and pathological features of the patients with IHs are shown in Table 2. The pathological diagnosis of the collected 5 pairs of IH samples, all of which were proliferative stage IH (Figure S1). FISH assays of the five pairs of IH samples and paraneoplastic tissues demonstrated that MIR4435-2HG expression was significantly higher in IH tissues, especially in the cytoplasm, than that in normal tissues (Figure 3a). qRT-PCR confirmed that MIR4435-2HG expression was higher in IH tissues than that in paraneoplastic tissues (Figure 3b). The subcellular localization of lncRNAs is closely related to their functions. Our results showed that MIR4435-2HG was mainly localized in the cytoplasm of HemECs (Figure 3c).
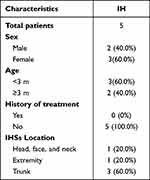 |
Table 2 Baseline Characteristics of Patients with IHs and Control Subjects |
In our further investigation of the biological role of MIR4435-2HG in HemECs, lentiviral vectors with MIR4435-2HG overexpression or knock down were constructed and successfully transfected into HemECs. qRT-PCR was used to verify the knockdown and overexpression efficiencies (Figure 3d). The CCK-8 assay showed that HemECs proliferation was diminished after MIR4435-2HG knockdown, whereas it was enhanced after MIR4435-2HG overexpression (Figure 3e). The wound healing assay showed that MIR4435-2HG knockdown in HemECs significantly inhibited wound healing, whereas MIR4435-2HG overexpression significantly promoted wound healing (Figure 3f and g). The Transwell invasion assay also showed that MIR4435-2HG knockdown significantly inhibited the invasion of HemECs, whereas MIR4435-2HG overexpression significantly promoted it (Figure 3h and i). Tube formation assays showed diminished tube formation capacity after MIR4435-2HG knockdown, with reductions in the number of junctions and meshes and total segments length; however, tube formation capacity was enhanced after MIR4435-2HG overexpression (Figure 3j–m).
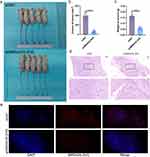
We established a subcutaneous tumor transplantation model to explore the role of MIR4435-2HG in vivo. The results showed that the size and weight of tumors after MIR4435-2HG knockdown were significantly lower than that in the shNC (Figure 4a–c), and the differences in the MIR4435-2HG overexpression group were not statistically significant (Figure S2a–c). Histological examination using hematoxylin and eosin (H&E) staining demonstrated that endothelial cell clumps aggregated and formed microvessels in the shNC group, whereas MIR4435-2HG knockdown was followed by an increase in fibrous tissue with only a few mature microvessels (Figure 4d). The FISH assay further demonstrated that MIR4435-2HG expression was associated with tumor growth (Figure 4e).
M2-Exos Transfer MIR4435-2HG from Macrophages to HemECs and Regulate the Function of HemECs
To further investigate whether MIR4435-2HG from M2-exos affects the function of HemECs, we added M2-exos to the medium containing both shNC and shMIR4435-2HG. The qRT-PCR assay showed that MIR4435-2HG expression was significantly increased after the addition of M2-exos (Figure 5a). The addition of M2-exos increased MIR4435-2HG expression level in shMIR4435-2HG to some extent. Moreover, the addition of M2-exos reversed proliferation (Figure 5b), migration (Figure 5c and d), invasion (Figure 5e and f), and angiogenesis (Figure 5g and j) of HemECs to some extent. Therefore, we hypothesized that M2-exos transfer MIR4435-2HG from macrophages to HemECs and affect their biological functions.
RNA-Seq Analysis After MIR4435-2HG Knockdown
We performed RNA-seq analysis of cells with MIR4435-2HG knockdown (GSE236203). A total of 3723 genes with statistically significant differences in expression were identified: 1636 upregulated and 2087 downregulated. Hierarchical clustering and volcano plots (Figure 6a and b) revealed that these genes were differentially expressed. Logarithmic fold change (|log2FC| ≥ 1 and p-adjusted <0.05) was used as the screening condition. To further investigate the functions of these target genes, we performed GO analysis and KEGG analysis for differentially expressed RNAs using the DAVID and KOBAS software. The top 20 significant pathways in the three components of GO analysis (biological process, molecular function, cellular component) (Figure S3a–c) included regulation of cellular processes, binding, and protein binding. KEGG analysis revealed the top 20 pathways associated with differentially expressed RNAs (Figure S3d), including the PI3K-Akt signaling pathway, focal adhesion, and ECM-receptor interaction. Our GSEA analysis showed that the NF-κB signaling pathway was significantly downregulated (Figure 6c). Western blotting detected the expression of NF-κB p65, p-p65, IκBα, and p-IκBα of the NF-κB signaling pathway. The expression of p-p65 and p-IκBα was significantly reduced after MIR4435-2HG knockdown (Figure 6d) and increased after MIR4435-2HG overexpression (Figure 6e).
MIR4435-2HG Affects RNA Expression of HNRNPA1 and Binds Directly to HNRNPA1 Protein
We determined the direct regulatory mechanisms of MIR4435-2HG. First, we synthesized an exogenous plasmid containing MIR4435-2HG or its antisense sequence (Figure S3e). Then, we linearized the RNA sequence and transcribed it (Figure S3f) in vitro. Biotin-labeled RNA was inoculated into protein lysates from HemECs, and both protein solutions were analyzed using mass spectrometry (Figure 6f). We combined the results of the previous RNA-seq analysis and unexpectedly found a decrease in the RNA expression of HNRNPA1 in the bind protein after MIR4435-2HG knockdown. We then used Western blotting and the product of another independent RNA pull-down experiment to validate the MIR4435-2HG-HNRNPA1 interactions (Figure 6g). RIP was performed in HemECs using the HNRNPA1 antibody. The MIR4435-2HG level in the HNRNPA1 RIP samples was much higher than that in the control IgG RIP samples (Figure 6h). HNRNPA1 expression was also decreased or increased in HemECs with MIR4435-2HG knockdown or overexpression, respectively, as confirmed by qRT-PCR (Figure 6i) and Western blotting (Figure 6j and k). This result suggests that MIR4435-2HG may be involved in the regulation of HNRNPA1 mRNA levels, consequently affecting HNRNPA1 expression.
HNRNPA1 is Regulated by MIR4435-2HG to Affect the Function of HemECs
We examined HNRNPA1 expression in five IH tissues and matched normal tissues using IHC staining. HNRNPA1 expression was significantly higher in IH tissues than that in normal tissues (Figure 7a and b). To explore whether HNRNPA1 also plays an influential role in HemECs, we constructed lentiviral vectors with of HNRNPA1 overexpression and knockdown and successfully transfected them into HemECs. qRT-PCR (Figure S4a) and Western blotting (Figure S4b and c) verified the knockdown and overexpression efficiency. The CCK-8 assay showed that the proliferation capacity of HemECs was reduced after HNRNPA1 knockdown, but was enhanced after HNRNPA1 overexpression (Figure 7c). The wound healing assay showed that cell migration ability was reduced after HNRNPA1 knockdown, but was enhanced after HNRNPA1 overexpression (Figure 7d and e). Transwell invasion analysis showed that the invasion ability of HemECs was reduced after HNRNPA1 knockdown, but was enhanced after HNRNPA1 overexpression (Figure 7f and g). Tube formation analysis showed that HNRNPA1 knockdown decreased tube formation ability with reductions in the number of junctions and meshes and total segments length; however, tube formation ability was enhanced after HNRNPA1 overexpression (Figure 7h–k).
To further investigate whether MIR4435-2HG affects the function of HemECs through HNRNPA1, we transfected shMIR4435-2HG to overexpress HNRNPA1 and an overexpression control virus. Western blotting showed that after transfection with the overexpression virus of HNRNPA1, shMIR4435- 2HG showed a remarkably increased HNRNPA1 expression (Figure 7l). Moreover, the effects MIR4435-2HG knockdown on the proliferation (Figure S5a), migration (Figure S5b and c), invasion (Figure S5d and e), and angiogenesis (Figure S5f–i) of HemECs was reversed to some extent after transfection with HNRNPA1 overexpression virus. Additionally, the Western blotting assay showed that HNRNPA1 overexpression reversed the effect on the NF-κB signaling pathway after MIR4435-2HG knockdown (Figure 7l).
Discussion
Despite being the most commonly observed vascular-derived tumor in infancy and childhood, the etiology, pathogenesis, and drug resistance of IHs remain unclear. M2-polarized macrophages are prevalent in proliferating IHs and are significantly reduced in number in involuting IHs.11 However, the role and mechanism of M2-polarized macrophages in the proliferation of IHs remain unclear. In this study, M2-exos-derived MIR44435-2HG enhanced the proliferation, migration, invasion, and angiogenesis of HemECs. Further experiments revealed that MIR44435-2HG affected HNRNPA1 expression and binds directly to HNRNPA1 to influence the NF-κB signaling pathway. This finding may help explain the effect of M2-polarized macrophages on IHs and suggests that MIR44435-2HG may be a new candidate oncogene in IHs.
M2 macrophages mediate tumor proliferation, invasion, metastasis, and angiogenesis through the release of exosomes. Cheng et al showed that M2-exos promoted HUVEC proliferation and inhibited HUVEC inflammation and apoptosis.24 Wei et al showed that M2-exos containing miRNAs promoted lung adenocarcinoma cell invasion, migration, and angiogenesis.25 Yang et al found that M2-exos inhibited E2F2 expression in endothelial cells, thereby promoting angiogenesis.26 Although research in on IHs is nascent, the available results are exciting. M2-polarized macrophages may promote the progression of IHs by facilitating angiogenesis; however, M1-polarized (not M2-polarized) macrophages induce endothelial-mesenchymal transition in HemECs and promote the regression of IHs.27 Liu et al found that M2-exos reduced the sensitivity of HemSCs to propranolol.14 These studies suggest a positive role of M2-exos in the pathophysiological development of IHs. Our study further demonstrated that M2-exos promotes proliferation, invasion, migration, and angiogenesis of HemECs while transporting MIR44435-2HG to influence the progression of IHs. These findings improve the understanding of the interactions between macrophage-derived exosomes and HemECs and broaden the understanding of the progression of IHs.
LncRNAs are widely present in organisms and are involved in the regulation of IHs. LncRNA-TUG1 promotes the proliferation, migration, and invasion of HemECs by regulating the miR-137/IGFBP5 axis.28 Silencing MALAT1 significantly inhibits the proliferation, migration, and angiogenesis of HemECs, but promotes apoptosis.29 MIR4435- 2HG participates in the progression of several human diseases and interacts with DNA, RNA, or proteins to regulate transcription, epigenetic modifications, protein/RNA stability, translation, and post-translational modifications.30 Cell proliferation, EMT, invasion, migration, and apoptosis inhibition are the main phenotypes of cancers that are affected by MIR4435-2HG expression.31 MIR4435-2HG expression was higher in IH tissues than that in paraneoplastic tissues. MIR4435-2HG was closely associated with the biological functions of HemECs, and MIR4435-2HG knockdown inhibited the growth of IHs in vivo. Unfortunately, the results of the in vivo tumorigenic experiments with MIR4435-2HG overexpression were not statistically different, which may be related to the higher native MIR4435-2HG expression in IHs. Thus, MIR4435-2HG is expected to be a novel target for IH treatment.
We used RNA-seq to explore the mechanism of MIR4435-2HG and found that the RNA expression of HNRNPA1 was regulated by MIR4435-2HG. Using RNA pull-down experiments, we found that MIR4435-2HG directly binds to HNRNPA1. HNRNPA1 is one of the most abundant RNA-binding proteins in the hnRNP family, and its key functions in tumors are particularly evident.32 It is involved in the key hallmarks of cancer, such as increased cell growth, survival, metastasis, modified cellular energetics, and immortalization.33 Multiple lncRNAs have been identified to regulate disease progression by binding to HNRNPA1. For example, the interaction between lncRNA SNHG6 and HNRNPA1 promotes colorectal cancer growth,34 and lncRNA ANCR promotes hepatocellular carcinoma metastasis by binding to HNRNPA1 and upregulating its expression.35 In this study, we confirmed that HNRNPA1 is highly expressed in IHs and regulates the biological functions of HemECs. Moreover, MIR4435-2HG can act on the NF-κB signaling pathway through HNRNPA1. The NF-κB signaling pathway can promote tumor angiogenesis.36,37 The NF-κB signaling pathway is highly expressed in proliferating IHs38,39 and HNRNPA1 is involved in regulating NF-κB-dependent transcription.40 Our results corroborate and enrich these findings.
Although we experimentally demonstrated that M2-exos regulate the development of IHs through the MIR4435-2HG/HNRNPA1/NF-κB axis, we also encountered some confusion. Macrophages in IHs originate from and in conditions under which they are transformed into M1- and M2-polarized macrophages. By contrast, lncRNAs localized in the nucleus can interact with various protein molecules, such as transcription factors, to regulate chromosome structure and function.41 lncRNAs localized in the cytoplasm with binding proteins mostly regulate mechanisms, such as mRNA variable shear, stability, and protein modifications. We demonstrated that MIR4435-2HG and HNRNPA1 interact to regulate the phenotype of IHs; however, how they bind and their function remains unclear.
Our study has some limitations. First, although we explored the effects of M2-polarized macrophage-derived exosomes on HemECs in vitro, we did not validate our results through vivo experiments. Second, because oral propranolol has become the first-line treatment option for IH, the tissue sample size was too small to determine whether MIR4435-2HG and HNRNPA1 can be used as biomarkers. Third, although regulation of the NF-κB signaling pathway was not the focus of this study, the exact point at which it intervenes in regulation remains to be explored in depth. These limitations provide topics for further research.
Conclusion
Our results suggest that M2-exos-derived MIR4435-2HG regulates the proliferation, migration, invasion, and angiogenesis of HemECs by binding to the HNRNPA1/NF-κB axis. Our findings complement the literature on the pathogenesis of IHs and may lead to the development of targeted exosome-related therapies.
Abbreviations
CCK-8, cell counting kit-8; ceRNA, competitive endogenous RNA network; H&E, hematoxylin and eosin; HemECs, hemangioma-derived endothelial cells; HNRNPA1, heterogeneous nuclear ribonucleoprotein A1; IH, infantile hemangioma; IHC, immunohistochemical; FISH, fluorescence in situ hybridization; M2-exos, M2 polarized macrophage-derived exosomes; PBS, phosphate buffered saline; RIP, RNA immunoprecipitation; TEM, transmission electron microscopy.
Acknowledgments
We would like to thank Editage for English language editing. And we would like to thank Shizhe Liu for his technical assistance.
Funding
Gratefully acknowledges the Shandong Natural Science Foundation (ZR2022MH080) the Clinical medical science innovation program of Jinan (202019076) and Taishan Scholars(No.ts201511100) to support the work of this study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Léauté-Labrèze C, Harper JI, Hoeger PH. Infantile haemangioma. Lancet. 2017;390(10089):85–94. doi:10.1016/S0140-6736(16)00645-0
2. Schrenk S, Boscolo E. A transcription factor is the target of propranolol treatment in infantile hemangioma. J Clin Invest. 2022;132(3):e156863. doi:10.1172/JCI156863
3. Chen J, Chen Q, Qiu Y, et al. CD146+ mural cells from infantile hemangioma display proangiogenic ability and adipogenesis potential in vitro and in xenograft models. Front Oncol. 2023;13:1063673. doi:10.3389/fonc.2023.1063673
4. Fu R, Zou Y, Wu Z, et al. Safety of oral propranolol for neonates with problematic infantile hemangioma: a retrospective study in an Asian population. Sci Rep. 2023;13(1):5956. doi:10.1038/s41598-023-33105-2
5. Kong M, Li Y, Wang K, Zhang S, Ji Y. Infantile hemangioma models: is the needle in a haystack? J Transl Med. 2023;21(1):308. doi:10.1186/s12967-023-04144-0
6. Léauté-Labrèze C, Hoeger P, Mazereeuw-Hautier J, et al. A randomized, controlled trial of oral propranolol in infantile hemangioma. N Engl J Med. 2015;372(8):735–746. doi:10.1056/NEJMoa1404710
7. Gong X, Li Y, Yang K, Chen S, Ji Y. Infantile hepatic hemangiomas: looking backwards and forwards. Precis Clin Med. 2022;5(1):bac006. doi:10.1093/pcmedi/pbac006
8. Komal S, Han SN, Cui LG, et al. Epigenetic regulation of macrophage polarization in cardiovascular diseases. Pharmaceuticals. 2023;16(2):141. doi:10.3390/ph16020141
9. Wang H, Ye X, Spanos M, et al. Exosomal non-coding RNA mediates macrophage polarization: roles in cardiovascular diseases. Biology. 2023;12(5):745. doi:10.3390/biology12050745
10. Zhang W, Chen G, Wang FQ, et al. Macrophages contribute to the progression of infantile hemangioma by regulating the proliferation and differentiation of hemangioma stem cells. J Invest Dermatol. 2015;135(12):3163–3172. doi:10.1038/jid.2015.321
11. Wang FQ, Chen G, Zhu JY, et al. M2-polarised macrophages in infantile haemangiomas: correlation with promoted angiogenesis. J Clin Pathol. 2013;66(12):1058–1064. doi:10.1136/jclinpath-2012-201286
12. Liao XM, Guan Z, Yang ZJ, et al. Comprehensive analysis of M2 macrophage-derived exosomes facilitating osteogenic differentiation of human periodontal ligament stem cells. BMC Oral Health. 2022;22(1):647. doi:10.1186/s12903-022-02682-5
13. Wang QZ, Zhao ZL, Liu C, Zheng JW. Exosome-derived miR-196b-5p facilitates intercellular interaction in infantile hemangioma via down-regulating CDKN1B. Ann Transl Med. 2021;9(5):394. doi:10.21037/atm-20-6456
14. Liu C, Zhao Z, Guo S, Zhang L, Fan X, Zheng J. Exosomal miR-27a-3p derived from tumor-associated macrophage suppresses propranolol sensitivity in infantile hemangioma. Cell Immunol. 2021;370:104442. doi:10.1016/j.cellimm.2021.104442
15. Hong Y, Zhang Y, Zhao H, Chen H, Yu QQ, Cui H. The roles of lncRNA functions and regulatory mechanisms in the diagnosis and treatment of hepatocellular carcinoma. Front Cell Dev Biol. 2022;10:1051306. doi:10.3389/fcell.2022.1051306
16. Sun CC, Li L, Jiang ZC, Liu ZC, Wang L, Wang HJ. The functional role of LncRNA UCA1 in pancreatic cancer: a mini-review. J Cancer. 2023;14(2):275–280. doi:10.7150/jca.79171
17. Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. 2021;21(7):446–460. doi:10.1038/s41568-021-00353-1
18. Zhong C, Xie Z, Zeng LH, Yuan C, Duan S. MIR4435-2HG is a potential pan-cancer biomarker for diagnosis and prognosis. Front Immunol. 2022;13:855078. doi:10.3389/fimmu.2022.855078
19. Li S, Hu X, Yu S, et al. Hepatic stellate cell-released CXCL1 aggravates HCC malignant behaviors through the MIR4435-2HG / miR -506-3p/ TGFB1 axis. Cancer Sci. 2023;114(2):504–520. doi:10.1111/cas.15605
20. Ke J, Wang Q, Zhang W, Ni S, Mei H. LncRNA MIR4435-2HG promotes proliferation, migration, invasion and epithelial mesenchymal transition via targeting miR-22-3p/TMEM9B in breast cancer. Am J Transl Res. 2022;14(8):5441–5454.
21. Yang T, Li Y, Wang G, et al. lncRNA MIR4435-2HG accelerates the development of bladder cancer through enhancing IQGAP3 and CDCA5 Expression. Biomed Res Int. 2022;2022:3858249. doi:10.1155/2022/3858249
22. Zhang M, Yu X, Zhang Q, Sun Z, He Y, Guo W. MIR4435-2HG: a newly proposed lncRNA in human cancer. Biomed Pharmacother. 2022;150:112971. doi:10.1016/j.biopha.2022.112971
23. Wang L, Zou Y, Huang Z, et al. KIAA1429 promotes infantile hemangioma regression by facilitating the stemness of hemangioma endothelial cells. Cancer Sci. 2023;114(4):1569–1581. doi:10.1111/cas.15708
24. Cheng X, Zhou H, Zhou Y, Song C. M2 macrophage-derived exosomes inhibit apoptosis of HUVEC cell through regulating miR-221-3p expression. Biomed Res Int. 2022;2022:1609244. doi:10.1155/2022/1609244
25. Wei K, Ma Z, Yang F, et al. M2 macrophage-derived exosomes promote lung adenocarcinoma progression by delivering miR-942. Cancer Lett. 2022;526:205–216. doi:10.1016/j.canlet.2021.10.045
26. Yang Y, Guo Z, Chen W, et al. M2 macrophage-derived exosomes promote angiogenesis and growth of pancreatic ductal adenocarcinoma by targeting E2F2. Mol Ther. 2021;29(3):1226–1238. doi:10.1016/j.ymthe.2020.11.024
27. Wu K, Muratore CS, So EY, et al. M1 macrophage-induced endothelial-to-mesenchymal transition promotes infantile hemangioma regression. Am J Pathol. 2017;187(9):2102–2111. doi:10.1016/j.ajpath.2017.05.014
28. Zhou L, Jia X, Yang X. LncRNA-TUG1 promotes the progression of infantile hemangioma by regulating miR-137/IGFBP5 axis. Hum Genomics. 2021;15(1):50. doi:10.1186/s40246-021-00349-w
29. Li MM, Dong CX, Sun B, et al. LncRNA-MALAT1 promotes tumorogenesis of infantile hemangioma by competitively binding miR-424 to stimulate MEKK3/NF-κB pathway. Life Sci. 2019;239:116946. doi:10.1016/j.lfs.2019.116946
30. Zhao F, Liu Y, Tan F, et al. MIR4435-2HG: a tumor-associated long non-coding RNA. Curr Pharm Des. 2022;28(25):2043–2051. doi:10.2174/1381612828666220607100228
31. Ghasemian M, Rajabibazl M, Sahebi U, et al. Long non-coding RNA MIR4435-2HG: a key molecule in progression of cancer and non-cancerous disorders. Cancer Cell Int. 2022;22(1):215. doi:10.1186/s12935-022-02633-8
32. Han P, Cao P, Yue J, et al. Knockdown of hnRNPA1 promotes NSCLC metastasis and EMT by regulating alternative splicing of LAS1L exon 9. Front Oncol. 2022;12:837248. doi:10.3389/fonc.2022.837248
33. Roy R, Huang Y, Seckl MJ, Pardo OE. Emerging roles of hnRNPA1 in modulating malignant transformation. Wiley Interdiscip Rev RNA. 2017;8(6). doi:10.1002/wrna.1431
34. Lan Z, Yao X, Sun K, Li A, Liu S, Wang X. The interaction between lncRNA SNHG6 and hnRNPA1 contributes to the growth of colorectal cancer by enhancing aerobic glycolysis through the regulation of alternative splicing of PKM. Front Oncol. 2020;10:363. doi:10.3389/fonc.2020.00363
35. Wen Z, Lian L, Ding H, et al. LncRNA ANCR promotes hepatocellular carcinoma metastasis through upregulating HNRNPA1 expression. RNA Biol. 2020;17(3):381–394. doi:10.1080/15476286.2019.1708547
36. Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–436. doi:10.1038/nature04870
37. Wang R, Ma Y, Zhan S, et al. B7-H3 promotes colorectal cancer angiogenesis through activating the NF-κB pathway to induce VEGFA expression. Cell Death Dis. 2020;11(1):55. doi:10.1038/s41419-020-2252-3
38. Greenberger S, Adini I, Boscolo E, Mulliken JB, Bischoff J. Targeting NF-κB in infantile hemangioma-derived stem cells reduces VEGF-A expression. Angiogenesis. 2010;13(4):327–335. doi:10.1007/s10456-010-9189-6
39. Xu W, Li S, Yu F, et al. Role of thrombospondin-1 and nuclear factor-κB signaling pathways in antiangiogenesis of infantile hemangioma. Plast Reconstr Surg. 2018;142(3):310e–321e. doi:10.1097/PRS.0000000000004684
40. Hay DC, Kemp GD, Dargemont C, Hay RT. Interaction between hnRNPA1 and IkappaBalpha is required for maximal activation of NF-kappaB-dependent transcription. Mol Cell Biol. 2001;21(10):3482–3490. doi:10.1128/MCB.21.10.3482-3490.2001
41. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi:10.1038/s41580-020-00315-9
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.