Back to Journals » International Journal of Nanomedicine » Volume 13
Lyophilization and stability of antibody-conjugated mesoporous silica nanoparticle with cationic polymer and PEG for siRNA delivery
Authors Ngamcherdtrakul W, Sangvanich T, Reda M, Gu S , Bejan D, Yantasee W
Received 2 February 2018
Accepted for publication 28 April 2018
Published 10 July 2018 Volume 2018:13 Pages 4015—4027
DOI https://doi.org/10.2147/IJN.S164393
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Worapol Ngamcherdtrakul,1,2,* Thanapon Sangvanich,1,* Moataz Reda,1 Shenda Gu,1 Daniel Bejan,2 Wassana Yantasee1,2
1Department of Biomedical Engineering, Oregon Health and Science University, Portland, OR, USA; 2Nanomedicine Research Unit, PDX Pharmaceuticals, LLC, Portland, OR, USA
*These authors contributed equally to this work
Introduction: Long-term stability of therapeutic candidates is necessary toward their clinical applications. For most nanoparticle systems formulated in aqueous solutions, lyophilization or freeze-drying is a common method to ensure long-term stability. While lyophilization of lipid, polymeric, or inorganic nanoparticles have been studied, little has been reported on lyophilization and stability of hybrid nanoparticle systems, consisting of polymers, inorganic particles, and antibody. Lyophilization of complex nanoparticle systems can be challenging with respect to preserving physicochemical properties and the biological activities of the materials. We recently reported an effective small-interfering RNA (siRNA) nanoparticle carrier consisting of 50-nm mesoporous silica nanoparticles decorated with a copolymer of polyethylenimine and polyethyleneglycol, and antibody.
Materials and methods: Toward future personalized medicine, the nanoparticle carriers were lyophilized alone and loaded with siRNA upon reconstitution by a few minutes of simple mixing in phosphate-buffered saline. Herein, we optimize the lyophilization of the nanoparticles in terms of buffers, lyoprotectants, reconstitution, and time and temperature of freezing and drying steps, and monitor the physical and chemical properties (reconstitution, hydrodynamic size, charge, and siRNA loading) and biological activities (gene silencing, cancer cell killing) of the materials after storing at various temperatures and times.
Results: The material was best formulated in Tris-HCl buffer with 5% w/w trehalose. Freezing step was performed at -55°C for 3 h, followed by a primary drying step at -40°C (100 μBar) for 24 h and a secondary drying step at 20°C (20 μBar) for 12 h. The lyophilized material can be stored stably for 2 months at 4°C and at least 6 months at -20°C.
Conclusion: We successfully developed the lyophilization process that should be applicable to other similar nanoparticle systems consisting of inorganic nanoparticle cores modified with cationic polymers, PEG, and antibodies.
Keywords: nanoparticles, lyophilization, cancer, mesoporous silica, antibody, siRNA
Introduction
In the last decade, nanoparticles have been widely developed as carriers for the delivery of antibodies, oligonucleotides, and drugs. Nanoparticles protect cargos against enzymatic degradation, prevent rapid clearance of small compounds by the kidneys, and prolong blood circulation half-life of the cargos.1 Typically, nanoparticles are formulated in solution as colloidal systems, which cannot be stored long term due to physical instability (aggregation) and chemical instability.2 To facilitate long-term storage, all traces of water must be removed by a process of freeze-drying or lyophilization.3,4 However, lyophilization of nanoparticles is more challenging than that of traditional chemical compounds since the process may affect both the physical (eg, size) and chemical properties of the nanoparticles. This is especially true when the nanoparticle consists of many components. Therefore, optimization of the lyophilization process and stability assessment are clearly needed.
We have recently developed cationic polymer modified mesoporous silica nanoparticles (MSNPs) as a promising small-interfering RNA (siRNA) carrier for breast cancer treatment.5 As shown in Figure 1, the MSNP of 50 nm in size was surface modified with a cross-linked polyethylenimine (PEI) and polyethyleneglycol (PEG). The cross-linked PEI allows the loading of negatively charged siRNA and promotes endosomal escape via proton sponge effect, while PEG provides a steric effect that protects siRNA from enzymatic degradation and nanoparticles from aggregation and phagocytosis. The PEI was cross-linked to increase the buffering capacity and enhance the endosomal escape of siRNA by the proton sponge effect principle. The modified nanoparticles are then conjugated with antibodies for targeting the cancer cells of interest. Specifically, Trastuzumab (Herceptin, Genentech), a humanized monoclonal HER2 antibody, was used as a homing agent for HER2-positive cancer. The siRNA nanoconstruct has been shown to overcome drug resistance in two HER2-positive cancer mouse models.5,6 Toward its clinical evaluation, material stability over a long period of time is needed.
  | Figure 1 Mesoporous silica-based nanoconstruct for targeted delivery of siRNA. |
Lyophilization of polymeric nanoparticles systems, such as poly(lactic-co-glycolic acid),7,8 polycaprolactone,9 or PEI,10 and lipid nanoparticle systems have been explored.11–14 Likewise, lyophilization of inorganic nanoparticle systems, such as silica15 or gold16 nanoparticles, has also been attempted. However, little has been reported on lyophilization and stability of hybrid nanoparticle systems, consisting of polymers, inorganic particles, and antibodies. These hybrid systems have been studied intensively for siRNA and drug delivery in the past several years due to the recognition that lipid or polymeric systems alone have not reached desired clinical outcomes, and hybrid systems, or at least targeting agents (eg, antibody), may be needed. Amine-modified silica nanoparticles were successfully lyophilized in the presence of trehalose (TL) as a lyoprotectant;15 however, the long lyophilization time of over 6 days is not highly economical.
Our aim was to achieve a stable long-term storage of the antibody (trastuzumab)-conjugated PEG-PEI-modified MSNP for siRNA delivery. Ideally, the material should be kept stable for over 6 months without requiring an expensive −80°C freezer. It must be easily reconstituted and maintain the properties and performance of the material for siRNA delivery. Trastuzumab has been lyophilized with TL to sustain protein structure and activity during long-term storage,17 but its lyophilization when conjugated on nanoparticles has not been reported. We adopt a two-vial strategy for our material (nanoparticles and siRNA were lyophilized separately) for its future use in personalized medicine, in which different siRNAs can be used on the nanoparticle carriers when oncogenes on the tumors are identified. This strategy exploits the rapid and simple loading of siRNA on our nanoparticle, which will be demonstrated.
Materials and methods
Chemicals and reagents
Tetraethyl orthosilicate (TEOS), cetyltrimethylammonium chloride (CTAC), sodium phosphate mono basic (NaH2PO4·H2O), sodium phosphate dibasic (Na2HPO4), triethanolamine (TEA), and TL dehydrate were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). Branched-PEI (10 kDa) was purchased from Alfa Aesar (Ward Hill, MA, USA). Maleimide-PEG (5 kDa)-NHS was purchased from JenKem Technology USA (Plano, TX, USA). Trastuzumab (Herceptin®, Genentech) was obtained from the OHSU pharmacy. Phosphate-buffered saline (PBS) (pH 7.2) was obtained from Life Technologies (Carlsbad, CA, USA). Zeba spin desalting columns (MW 40 kDa), RNase-free water, Traut’s reagent, dithiobis (succinimidyl propionate) (DSP), ethanol, HCl, sodium hydroxide, and DharmaFECT#1 were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Tris-HCl buffer was purchased from Boston BioProducts (Ashland, MA, USA). All reagents are of the highest purity grade available. RPMI-1640 cell media, fetal bovine serum (FBS), and penicillin/streptomycin (P/S) were purchased from Life Technologies.
siHER2 (sense: 5′ CACGUUUGAGUCCAUGCCCAAUU 3′, antisense: 5′ UUGGGCA-UGGACUCAAACGUGUU 3′), siSCR (sense: 5′ UGGUUUACAUGUCGACUAA 3′, anti-sense: 5′ UUAGUCGACAUGUAAACCA 3′), and DY677-siSCR were custom synthesized (in vivo HPLC grade) by Dharmacon and Thermo Fisher Scientific (Lafayette, CO, USA). siLUC (sense: 5′ CGGAUUACCAGGGAUUUCAtt 3′, antisense: 5′ UGAAAUCCCUGGUAAUCCGtt 3′) was custom synthesized by Life Technologies.
Nanoparticle synthesis
Nanoparticles were synthesized following our published protocol.5 Briefly, CTAC surfactant (0.15 M) was mixed with TEA (7 mL) in 2.5 L of water at 95°C. TEOS (60 mL) was then slowly added to the mixture under vigorous stirring for 1 h. The nanoparticles were then recovered by centrifugation, washed twice with ethanol, and dried overnight. The dried nanoparticles were resuspended and refluxed in acidic methanol (0.6 M HCl), recovered, washed with ethanol, and dried in a desiccator to obtain MSNPs. The dry MSNPs with the size of 50 nm were then mixed with branched-PEI in absolute ethanol at a mass ratio of 4:1 of MSNP per PEI. The mixture was shaken continuously for 3 h at room temperature, centrifuged, and resuspended in the ethanol solution containing PEI and 0.2 mg DSP as a cross-linker. The mixture was shaken for 40 min, then washed twice to remove excess PEI and DSP. Maleimide-PEG (5 kDa)-NHS was conjugated to MSNP-PEI at a mass ratio of 1:1 in PBS pH 7.2 under shaking condition for 2 h. The MSNP-PEI-PEG was washed twice with the PBS, resuspended, and kept in PBS.
Antibody, trastuzumab (T), was conjugated to MSNP-PEI-PEG via a thiol-maleimide reaction following our published recipe.5 First, trastuzumab was thiolated with Traut’s reagent in phosphate buffer pH 8.0 with 50-fold molar excess of reagent for 2 h and purified by Zeba spin column. Thiolated trastuzumab was mixed with MSNP-PEI-PEG at a mass ratio of 1:10 overnight at 4°C under continuous stirring. The conjugated MSNP-PEI-PEG (denoted T-NP) was washed with copious amount of PBS to remove excess antibody. T-NP was then stored in PBS pH 7.2 until use.
The loading of siRNA was achieved by mixing T-NP and siRNA (at a nanoparticle/siRNA mass ratio of 50) in PBS solution under rigorous shaking of 250 rpm for 2.5–30 min at room temperature. The thermal gravimetric analysis (TEM) image of MSNP cores and the schematic of surface modification are presented in Figure 1. The material contained 65.3 wt.% of MSNP, 13.5 wt.% of PEI, 18.2 wt.% of PEG (all by TGA analysis), and 3 wt.% of trastuzumab by BCA analysis as reported previously.5 The hydrodynamic size in PBS of the final construct with 2 wt.% siRNA was 113 ± 2.2 nm with narrow size distribution (PDI of 0.2) and zeta potential of 9.56 ± 0.13 mV in 10 mM NaCl, considered in a neutral range according to the NCI’s Nanotechnology Characterization Lab (NCL). The hydrodynamic size of about 110 nm of the final nanoparticles (after the polymer coating, antibody attachment, and siRNA loading on the 50 nm [TEM size] MSNP core) indicated that the particles were not aggregated but remained as individual particles owing to the dense layer of PEG. If aggregated, the size would increase more significantly (eg, from 32 nm of MSNP core [TEM size] to 305 nm [DLS size in water] of the PEI-coated MSNP).18
Characterization of size and zeta potential by DLS
Hydrodynamic diameter and zeta potential (charge) evaluations were performed with a Zetasizer Nano ZS (Malvern Instruments, Westborough, MA, USA). For size measurement, the 100 μg/mL of the material in 100 mM Tris-HCl buffer or PBS was used. The charge was measured in 10 mM NaCl using the same suspension conditions. The samples were loaded in appropriate cuvettes/capillary cells, equilibrated to 25°C before a minimum of 3 measurements were made.
Freeze-thaw of nanoparticles (T-NP)
Nanoparticles at 10 mg/mL in 100 mM Tris-HCl buffer (pH 7.4) or PBS with 0%–10% TL (%w/w of TL per nanoparticle) as a lyoprotectant were slowly frozen at −1°C/min from room temperature to −80°C using a CoolCell® LX Freezing Container (BioCision, San Rafael, CA, USA). The frozen materials were slowly thawed on ice to room temperature before use.
Lyophilization of nanoparticle (T-NP)
Nanoparticle (T-NP) was first lyophilized without siRNA as shown in Figure 1C. T-NP was lyophilized at 10 mg/mL in 100 mM Tris-HCl buffer pH 7.4 or PBS with 0%–25% TL (%w/w) as the lyoprotectant. Then 500 μL of each nanoparticle formula was lyophilized in a 2-mL glass vial in a BenchTop Freeze Dryer (SP Scientific VirTis AdVantage 2.0, Warminster, PA, USA). The “initial” lyophilization conditions were adapted from the work by Sameti et al on amine-modified silica nanoparticle, which took 6 days.15 Specifically, the formulation was slowly frozen at a shelf temperature of −55°C for 6 h. Primary drying was performed at a shelf temperature of −55°C and a pressure of 100 μbar for 24 h. The shelf temperature was then gradually increased to −40°C, and the pressure was reduced to 20 μbar for secondary drying for 54 h. After completion, vials were capped under vacuum with a built-in stoppering function. This process worked well at preserving the material but still took 4 days. To reduce the lyophilization time further, the conditions were optimized as follows: reducing the freezing time from 6 to 3 h (still at −55°C), increasing the primary drying temperature to −40°C (100 μbar), and increasing the secondary temperature to 20°C (20 μbar) while shortening the time to 12 h (“optimized” condition). Thermocouples were inserted into representative vials to monitor the product temperature throughout the lyophilization process.
Storage, reconstitution, and siRNA loading
Lyophilized nanoparticles were used immediately or stored at 4 different temperatures: −20°C, 4°C, 20°C, and 37°C, for a specified period of time (up to 6 months) prior to characterization and performance evaluation. Prior to use, the lyophilized material was reconstituted with 500 μL of RNase-free water to 10 mg/mL. The suspension was sonicated for 1 min. Size and charge were measured as previously described. For siRNA loading, the reconstituted nanoparticles were mixed with siRNA in PBS to achieve an NP/siRNA mass ratio of 50. The mixture was shaken at 250 rpm and at room temperature for 2.5–30 min, and the material was ready for transfection in cells. The size and charge of the nanoconstruct (post siRNA binding) was also measured. To characterize siRNA loading, (Dy677) siSCR was loaded on the nanoparticle as aforementioned. Then the suspension was centrifuged at 21,130×g for 30 min, and the fluorescent signal of (Dy677)siSCR in the supernatant was measured by Tecan Infinite M200; negligible signal was found for all materials, indicating complete siRNA binding.
Luciferase knockdown
The LM2-4luc+/H2N cell line (overexpressing luciferase and HER2), a gift from Prof. Robert Kerbel (University of Toronto), was used for the initial gene silencing efficacy assessment of the nanoparticles as previously reported.5 The cells were derived from MDA-MB-231 human breast cancer cell line19 available through ATCC. Cells were cultured in RPMI-1640 supplemented with 5% FBS and 1X P/S at 37°C in 5% CO2 atmosphere. Cells were seeded at 3,500 cells/well in a 96-well plate under cell medium without antibiotics for 24 h prior to treatment. Nanoparticles loaded with siLUC or siSCR at an NP/siRNA mass ratio of 50 were applied to each well at a fixed dose of 30 nM siRNA. After overnight incubation (~20 h), cells were washed once and replenished with complete media containing antibiotics. At 48 h post treatment, cells were lysed and analyzed for luciferase activity by the Luciferase Glow Assay Kit (Thermo Fisher Scientific) and protein concentration by BCA protein assay kit (Thermo Fisher Scientific), following the manufacturer’s protocols. Luciferase activity of the lysate was normalized with the corresponding protein concentration in the same well.
Cancer cell death
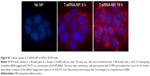
HER2+ human breast cancer cells, BT474, were obtained from ATCC. Cells were cultured in RPMI-1640 supplemented with 10% FBS. Cells were maintained at 37°C in 5% CO2 air atmosphere and were passaged weekly by trypsinization. Cells were plated in a 96-well plate with cell medium without antibiotics. One day after seeding, cells were treated with nanoparticles loaded with HER2 siRNA (siHER2) at an NP/siRNA ratio of 50. siRNA dose was 60 nM. The media was switched to complete media after overnight incubation. Figure S1 shows a significant cellular uptake of the nanoparticles within 1 h of exposure, and a significant siRNA transport within the cells at 6 and 16 h (higher signal intensity indicated endosomal escape of siRNA). Five days after treatment with T-siHER2-NP, cells were analyzed for viability using CellTiter-Glo® Luminescent Assay (Promega, Madison, WI, USA). The value was reported against scrambled siRNA counterpart (T-siSCR-NP).
Data and statistical analysis
Size and charge measurement were performed in triplicate. SiRNA binding was performed in duplicate. Gene silencing and cell viability were performed with 3–4 replicates. All data are reported as a mean ± SD. Comparisons of 2 groups were performed with Student’s t-tests (assuming normal distribution) using Statistical function of Excel. A p-value <0.05 was considered to be statistically significant.
Results and discussion
Buffer selection
To stabilize the pH, drugs or nanoparticles are prepared in physiological buffer systems such as PBS or Tris-HCl buffer. PBS was evaluated since our nanoparticle was synthesized, bound with siRNA, and initially kept in this buffer. Tris-HCl buffer was used because it was proven to be a bio-compatible buffer in lyophilizing silica nanoparticles.15 Its pH did not shift with freezing temperature20 unlike PBS shown in prior reports.20–22 We investigated the effect of both buffers on freeze-thawed and freeze-dried nanoparticles. The nanoparticles were suspended at 10 mg/mL with a TL content of 0% and 10% (w/w of nanoparticles) in either 1X PBS or 100 mM Tris-HCl (pH 7.4). The formulae were then frozen slowly at the rate of −1°C/min overnight. For the freeze-thaw study, the materials were thawed slowly back to room temperature. For the freeze-dry study, the materials underwent two drying steps as mentioned above. After reconstitution, hydrodynamic sizes of all freeze-thawed materials remained similar to that of the freshly made material as shown in Figure 2A. The freeze-thawed materials did not aggregate in PBS nor Tris-HCl buffer even without TL as a lyoprotectant. The lyophilized materials in PBS, however, aggregated even in the presence of TL (Figure 2B). The sizes increased to 1.5-fold over that of the freshly made counterpart. When lyophilized in Tris-HCl buffer, the particles did not aggregate and retained the original size when in the presence of 10% TL (Figure 2B). Furthermore, it was the only formulation that retained the size of the nanoconstruct once loaded with siRNA to that of the freshly made counterpart (Figure 2B). Although a previous report23 suggested that PBS may affect the freezing step (eg, drastic pH change due to a decrease in PBS solubility as temperature decreases) leading to unwanted particle aggregation, our data indicate that it was the drying step that caused our particle aggregation in the PBS system. However, low concentration of Tris-HCl buffer, which did not cause drastic pH change during the freezing step,20,24 appeared to protect the nanoparticle better in the drying step too. Thus, Tris-HCl was selected as a lyophilization buffer in all subsequent experiments.
Lyoprotectant selection and optimization
Lyoprotectant is a vital component in lyophilized nanoparticle formula. It protects nanoparticle stresses generated during the freezing and drying step of lyophilization.4 These stresses along with concentration changes encountered during the freezing step could cause particle aggregation, irreversible fusion, or destabilization.25 The most common lyoprotectants are sugars and other polyol compounds, such as TL, sucrose, glucose, sorbitol, and glycerol. We screened for the best lyoprotectants by performing a freeze-thaw experiment on our nanoparticles using all five lyoprotectants and found TL and sucrose to be the best at preserving the size and charge of our nanoparticles (not shown). However, since our nanoparticles are developed as siRNA carriers for cancer therapeutics, we chose TL over sucrose since there is a report that sucrose can accelerate tumor growth in tumor-bearing mice.26 TL was also previously reported as an effective lyoprotectant for silica nanoparticles,15 trastuzumab antibody,17 and siRNA on lipid nanoparticles.14
Next we investigated the concentration of TL required for lyophilization of our nanoparticles. We rejected the 0% TL condition since the material increased in size (Figure 2B). The nanoparticle suspension at 10 mg/mL in 100 mM Tris-HCl with a TL content of 5%–25% (%w/w of T-NP) were lyophilized under the aforementioned conditions. All lyophilized T-NP with a TL content of 5%–25% retained the average size and charge to that of the freshly made material after 1 min of sonication as shown in Figure 3A and B. However, with 25% TL, the finished product had partially collapsed cake, while lower TL contents produced perfect cakes (Figure S2). It is possible that high amount of TL caused a drop in collapse temperature of the mixture (ie, −30°C for TL–water binary mixture).27 When the product temperature during primary drying exceeds the collapse temperature, it causes loss of cake structure.28 Therefore, we rejected the 25% TL condition.
In addition to size and charge, luciferase knockdown and cancer cell killing were used to test the performance of the lyophilized materials upon loading with siLUC and siHER2, respectively, while siSCR was used as a negative control. When delivering siLUC, the lyophilized materials yielded comparable luciferase knockdown efficacy with freshly made material counterparts (from the same batch), but the 5% TL condition yielded the closest outcome (Figure 3C). When delivering siHER2, they also yielded comparable cell viability of BT474 cells with freshly made materials (Figure 3D). It is worth noting that the nanoconstruct contains trastuzumab, and thus killed about 50% of BT474, which is a very trastuzumab-sensitive cell line, as shown in our prior publication.5 We also showed that this killing effect was much lower in trastuzumab-resistant cell line (BT474 made resistant to trasutuzumab).5 Some exception was found with 0% TL condition, which yielded the material that was more toxic to cells than freshly made material (eg, greater nonspecific cell death with T-siSCR-NP). The two fold increase in size (see Figure 2B) may contribute to higher toxicity due to higher cellular uptake by mass. On the basis of the particle size and efficacy, the 5%–10% TL possessed similar characteristics as the freshly made counterpart, and the 5% TL was slightly better than the 10% TL condition based on the silencing efficacy. Since the lower amount of additive is more desirable for human applications, 5% TL was selected for subsequent lyophilization processes and long-term storage study.
Reconstitution
The working drug formula must be reconstituted easily in clinics. We tested the reconstitution of the optimal lyophilized T-NP (with 5% TL). Five hundred microliters of RNAse-free water was added slowly to the lyophilized cake. The suspension was vortexed for a given time and subjected to hydrodynamic size measurement. After 30 seconds of vortexing, the size was about 6 times of that of freshly made material and only reduced to about 3 times after a prolonged period of 10 min (Figure S3A). However, after only 1 min of sonication, the size and size distribution (Figure S3A and B) were the same with those of the freshly made material. Vigorous stirring could not bring the size down, suggesting particle agglomeration post lyophilization. A short sonication was needed to separate individual particles from each other. We concluded that 1 min of sonication was best for reconstitution of lyophilized T-NP and was used throughout the studies.
Time and temperature optimization
The initial lyophilization employed −55°C for 6 h during the freezing step, followed by 2 drying steps at −55°C for 24 h and −40°C for 54 h. The entire lyophilization process took almost 4 days. We proceeded to optimize the freezing time and drying temperature in order to shorten the entire process while preserving the characteristics and performance of the lyophilized materials. We started with the formulation of 10 mg/mL T-NP in 0.1 M Tris-HCl with 5%w/w TL. Freezing time was reduced from 6 to 3 h. During the primary drying, the product temperature must be above the collapse temperature to avoid collapsing of lyophilized cake.4 Primary drying temperature was increased to −40°C with a chamber pressure of 100 μbar. At this condition, the product temperature, monitored with thermocouple, was stabilized at −20°C for at least 10 h (overnight) before the primary drying was stopped to ensure complete ice sublimation. The total primary drying time was 24 h. Trastuzumab antibody was shown to preserve its protein structure after undergoing lyophilization with the secondary drying temperature of 20°C.17 Thus, we elevated the secondary drying temperature of our material (containing trastuzumab) from −40°C to 20°C under the chamber pressure of 20 μbar, which allowed us to shorten the drying time to 12 h. The lyophilized product under the “optimized” conditions demonstrated noncollapsed cake-like structure similar to Figure S2 (with 5% TL). At these new conditions, reconstitution condition, size, luciferase silencing, and cancer cell killing of the lyophilized T-NP were preserved similar to those from the original conditions and to those of freshly made materials (Figure 4), while the total time was reduced from 88 to 42 h. Hence, these optimized lyophilization conditions were used in the subsequent shelf-life study.
Long-term storage of lyophilized nanoparticles (T-NP)
To optimize the storage condition, the nanoparticles lyophilized under the aforementioned conditions were stored at 4 different temperatures: −20°C, 4°C, 20°C, and 37°C. The lyophilized products were evaluated bimonthly for physical appearance, size, charge, siRNA loading, luciferase silencing efficacy, and cancer cell killing efficacy. All materials (stored at 4 temperatures) retained the same cake appearance to the freshly lyophilized product. The materials were reconstituted and measured for hydrodynamic size (relative to freshly made material) as shown in Figure 5A. The material stored at 20°C started to aggregate at 6 weeks and the one stored at 37°C started to aggregate as soon as 2 weeks (ie, not fully reconstituted even after 7 min of sonication) (Figure 5A). The lyophilized products stored at −20°C and 4°C for up to 8 weeks were reconstituted effectively within 1 min of sonication to achieve the same size and size distribution of freshly made material (Figure 5B). At week 12, the material stored at 4°C had larger size distribution (Figure 5C), and the size was increased by 150% by week 16 of storage (Figure 5A). On the contrary, the material stored at −20°C continued to retain both the size and size distribution for at least 6 months with minimal changes (Figure 5D; longer term was not monitored).
On the basis of sizes, the best two storage temperatures were −20°C and 4°C. Next, we evaluated charge (zeta potential), siRNA loading, luciferase silencing efficacy, and cancer cell killing efficacy of lyophilized materials stored at both temperatures. The charge (Figure 6A) and siRNA loading (Figure 6B) of both materials remained similar to the freshly made nanoparticle. Luciferase silencing efficacy (Figure 6C; with siLUC) and cancer cell killing efficacy (reported as the viability of BT474 cells, Figure 6D, with siHER2) of both materials were also comparable to (or with marginal changes) the freshly made materials for up to 8 weeks, while the one stored at 4°C started to deviate from the performance of freshly made materials at 12 weeks. This was in agreement with the larger size and size distribution of the material (Figure 5C). Larger particle sizes lead to higher silencing efficacy and BT474 cell killing in vitro, but are not desirable for in vivo use. However, the material stored at −20°C retained comparable size (Figure 5A and D) and efficacies to those of freshly made materials (Figure 6C and D). We conclude that −20°C is the most suitable temperature for long-term storage of the nanoparticles.
  | Figure 6 Charge, siRNA loading, luciferase silencing efficacy, and cancer cell killing of lyophilized nanoparticles stored at various times and temperatures. |
In short, MSNP nanoparticles coated with PEI and PEG and conjugated with trastuzumab could be lyophilized and stored stably under −20°C for at least 6 months (longer term was not monitored). Lyophilization of siRNA is routinely done by vendors such as QIAGEN (Germantown, MD, USA) and can be kept stably at −20°C for 12 months according to QIAGEN; thus, it was not a subject of this work. Two vial approaches (ie, siRNA and nanoparticle carrier in separate vials) are preferred in a personalized medicine setting since it permits interchangeable siRNAs for targeting different genes using the same nanoparticle formulation. This is especially true for nanoparticle systems like ours, which allow easy loading of siRNA that can be done in clinics. Figure 7 shows that after just 2.5 min of binding in PBS, the T-NP and siRNA construct yielded comparable hydrodynamic size and luciferase silencing efficacy to those after 5, 10, and 30 min of binding. More conveniently, we can also add powders of siRNA and nanoparticle lyophilized separately into the same vial, and let them bind together upon reconstitution. This should help simplify the expensive stability studies of our materials during the IND enabling studies. We foresee no issue with this approach since both components are highly soluble in saline and achieve complete binding within a few minutes (Figure 7).
Conclusion
In summary, we successfully developed the lyophilization process for a hybrid polymer-inorganic nanoparticle system using a much shorter time than that reported for amine-modified MSNP (2 vs 6 days).15 The antibody-conjugated PEG-PEI-silica nanoparticles were lyophilized in Tris buffer with 5% TL as the lyoprotectant. The optimized conditions produced lyophilized material with cake-like structure and retained hydrodynamic size, charge (zeta potential), siRNA loading ability, silencing efficacy, and cancer cell killing efficacy to that of the freshly made material. The freeze-dried nanoparticles can be stored at −20°C for at least 6 months. Longer term stability (up to 2 years) will be evaluated by a Good Manufacturing Practice-certified contract research organization as we prepare the material for clinical trials using the lyophilization and storage conditions optimized herein. The sol–gel MSNP synthesis and the layer-by-layer modification of PEI, PEG, antibody, and siRNA on MSNPs offer good synthesis reproducibility and scalability. We have scaled up the synthesis protocol to yield 6 g of MSNP, which is 50-fold higher than our small-scale synthesis (human dose is anticipated to be 140–350 mg NP per dose). MSNPs have the same size and morphology for both small and large-scale syntheses (Figure S4). We have also reported the outstanding reproducibility of the nanoconstruct synthesis in terms of size (relative standard deviation [RSD; deviation from the mean] of 3.2% for 6 synthesis batches) and silencing efficacy (RSD of 2.3% from 6 batches).5 The lyophilization conditions, storage conditions, and material evaluation should be applicable to other similar nanoparticle systems consisting of inorganic nanoparticle cores that are surface modified with cationic polymers and PEG and conjugated with biomolecules like antibodies.
Acknowledgments
This work was supported by NCI/SBIR (Contract No HHSN261201300078C), NCI/SBIR (Grant No R44CA217534), NCATS/SBIR (Grant No R43TR001906), the Prospect Creek Foundation, and OHSU’s VPR fund. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors are grateful to Dr Robert Kerbel of University of Toronto for breast cancer cell lines, Samuel Mihelic and Brandon Beckman for their contribution in material synthesis and tissue culture work, and Dr Leslie Muldoon for her independent review of the data in this paper as required by OHSU conflict of interest guidelines.
Disclosure
OHSU, WN, and WY have a significant financial interest in PDX Pharmaceuticals, LLC, a company that may have a commercial interest in the results of this research and technology. This potential personal and institutional conflict of interest has been reviewed and managed by OHSU. The authors report no other conflicts of interest in this work.
References
Ngamcherdtrakul W, Castro DJ, Gu S, et al. Current development of targeted oligonucleotide-based cancer therapies: perspective on HER2-positive breast cancer treatment. Cancer Treat Rev. 2016;45:19–29. | ||
Chacón M, Molpeceres J, Berges L, Guzmán M, Aberturas MR. Stability and freeze-drying of cyclosporine loaded poly(D,L-lactide-glycolide) carriers. Eur J Pharm Sci. 1999;8(2):99–107. | ||
Franks F. Freeze-drying of bioproducts: putting principles into practice. Eur J Pharm Biopharm. 1998;45(3):221–229. | ||
Abdelwahed W, Degobert G, Stainmesse S, Fessi H. Freeze-drying of nanoparticles: formulation, process and storage considerations. Adv Drug Deliv Rev. 2006;58(15):1688–1713. | ||
Ngamcherdtrakul W, Morry J, Gu S, et al. Cationic polymer modified mesoporous silica nanoparticles for targeted siRNA delivery to HER2+ breast cancer. Adv Func Mater. 2015;25(18):2646–2659. | ||
Gu S, Hu Z, Ngamcherdtrakul W, et al. Therapeutic siRNA for drug-resistant HER2-positive breast cancer. Oncotarget. 2016;7(12):14727–14741. | ||
Holzer M, Vogel V, Mantele W, Schwartz D, Haase W, Langer K. Physico-chemical characterisation of PLGA nanoparticles after freeze-drying and storage. Eur J Pharm Biopharm. 2009;72(2):428–437. | ||
Fonte P, Araujo F, Seabra V, Reis S, van de Weert M, Sarmento B. Co-encapsulation of lyoprotectants improves the stability of protein-loaded PLGA nanoparticles upon lyophilization. Int J Pharm. 2015;496(2):850–862. | ||
Saez A, Guzman M, Molpeceres J, Aberturas MR. Freeze-drying of polycaprolactone and poly(D,L-lactic-glycolic) nanoparticles induce minor particle size changes affecting the oral pharmacokinetics of loaded drugs. Eur J Pharm Biopharm. 2000;50(3):379–387. | ||
Brus C, Kleemann E, Aigner A, Czubayko F, Kissel T. Stabilization of oligonucleotide-polyethylenimine complexes by freeze-drying: physicochemical and biological characterization. J Control Release. 2004;95(1):119–131. | ||
Yadava P, Gibbs M, Castro C, Hughes JA. Effect of lyophilization and freeze-thawing on the stability of siRNA-liposome complexes. AAPS PharmSciTech. 2008;9(2):335–341. | ||
Maitani Y, Aso Y, Yamada A, Yoshioka S. Effect of sugars on storage stability of lyophilized liposome/DNA complexes with high transfection efficiency. Int J Pharm. 2008;356(1–2):69–75. | ||
Kundu AK, Chandra PK, Hazari S, et al. Stability of lyophilized siRNA nanosome formulations. Int J Pharm. 2012;423(2):525–534. | ||
Ball RL, Bajaj P, Whitehead KA. Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization. Int J Nanomedicine. 2017;12:305–315. | ||
Sameti M, Bohr G, Ravi Kumar MN, et al. Stabilisation by freeze-drying of cationically modified silica nanoparticles for gene delivery. Int J Pharm. 2003;266(1–2):51–60. | ||
Alkilany AM, Abulateefeh SR, Mills KK, et al. Colloidal stability of citrate and mercaptoacetic acid capped gold nanoparticles upon lyophilization: effect of capping ligand attachment and type of cryoprotectants. Langmuir. 2014;30(46):13799–13808. | ||
Cleland JL, Lam X, Kendrick B, et al. A specific molar ratio of stabilizer to protein is required for storage stability of a lyophilized monoclonal antibody. J Pharm Sci. 2001;90(3):310–321. | ||
Buchman YK, Lellouche E, Zigdon S, Bechor M, Michaeli S, Lellouche JP. Silica nanoparticles and polyethyleneimine (PEI)-mediated functionalization: a new method of PEI covalent attachment for siRNA delivery applications. Bioconjug Chem. 2013;24(12):2076–2087. | ||
du Manoir JM, Francia G, Man S, et al. Strategies for Delaying or Treating In vivo Acquired Resistance to Trastuzumab in Human Breast Cancer Xenografts. Clin Cancer Res. 2006;12(3):904–916. | ||
Kolhe P, Amend E, Singh SK. Impact of freezing on pH of buffered solutions and consequences for monoclonal antibody aggregation. Biotechnol Prog. 2010;26(3):727–733. | ||
Pikal-Cleland KA, Carpenter JF. Lyophilization-induced protein denaturation in phosphate buffer systems: monomeric and tetrameric beta-galactosidase. J Pharm Sci. 2001;90(9):1255–1268. | ||
Croyle MA, Cheng X, Wilson JM. Development of formulations that enhance physical stability of viral vectors for gene therapy. Gene Ther. 2001;8(17):1281–1290. | ||
Pikal-Cleland KA, Rodriguez-Hornedo N, Amidon GL, Carpenter JF. Protein denaturation during freezing and thawing in phosphate buffer systems: monomeric and tetrameric beta-galactosidase. Arch Biochem Biophys. 2000;384(2):398–406. | ||
Carpenter JF, Pikal MJ, Chang BS, Randolph TW. Rational design of stable lyophilized protein formulations: some practical advice. Pharm Res. 1997;14(8):969–975. | ||
Abdelwahed W, Degobert G, Fessi H. A pilot study of freeze drying of poly(epsilon-caprolactone) nanocapsules stabilized by poly(vinyl alcohol): formulation and process optimization. Int J Pharm. 2006;309(1–2):178–188. | ||
Kimura Y, Sumiyoshi M. High-fat, high-sucrose, and high-cholesterol diets accelerate tumor growth and metastasis in tumor-bearing mice. Nutr Cancer. 2007;59(2):207–216. | ||
Yang G, Gilstrap K, Zhang A, Xu LX, He X. Collapse temperature of solutions important for lyopreservation of living cells at ambient temperature. Biotechnol Bioeng. 2010;106(2):247–259. | ||
Patel SM, Doen T, Pikal MJ. Determination of end point of primary drying in freeze-drying process control. AAPS PharmSciTech. 2010;11(1):73–84. |
Supplementary materials
  | Figure S2 Products from lyophilization process with 5 mg of nanoparticle (T-NP) and 0%–25% w/w TL. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.