Back to Journals » OncoTargets and Therapy » Volume 9
Lrig1 is a positive prognostic marker in hepatocellular carcinoma
Authors Yang B, Dai C, Tan RM, Zhang B, Meng X, Ye J, Wang XQ, Wei L, He F, Chen ZS
Received 11 May 2016
Accepted for publication 17 September 2016
Published 15 November 2016 Volume 2016:9 Pages 7071—7079
DOI https://doi.org/10.2147/OTT.S112534
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Bo Yang,1,2 Chen Dai,1,2 Rumeng Tan,1,2 Bo Zhang,1,2 Xiao Meng,3 Jing Ye,3 Xinqiang Wang,1,2 Lai Wei,1,2 Fan He,4 Zhishui Chen1,2
1Institute of Organ Transplantation, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, People’s Republic of China; 2Key Laboratory of Ministry of Health and Key Laboratory of Ministry of Education, Wuhan, Hubei, People’s Republic of China; 3Department of Pathology, Liaocheng People’s Hospital, Liaocheng, Shandong, People’s Republic of China; 4Department of Nephrology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, People’s Republic of China
Background: The prevalence of hepatocellular carcinoma (HCC) is increasing worldwide. As a consequence, there is an urgent need for identifying molecular markers of HCC development and progression. Recently, several studies have suggested that the Lrig1 may have prognostic implications in various cancer types, but its clinical value in HCC is not well evaluated.
Materials and methods: In this study, the expression level of Lrig1 was examined in 133 HCC tissues and adjacent normal tissues by immunohistochemistry. Furthermore, potential associations between Lrig1 expression and the carcinoma clinical parameters were investigated, including recurrence and survival rate. We silenced the Lrig1 in the normal liver cell line (LO2) and liver cancer cell line (Hep-G2) in vitro by the small interference RNA and detected its influence on proliferation, migration, and invasion.
Results: The expression of Lrig1 was significantly downregulated in liver cancer tissues and cell lines, and its expression levels were related to tumor size, tumor–node–metastasis staging and tumor recurrence. Furthermore, analysis of 6-year survival of 133 HCC patients showed that those with stronger Lrig1 expression had significantly longer overall survival time than those with weaker Lrig1 expression. In addition, decreased expression of Lrig1 in vitro promoted the growth, migration, or invasion of normal liver cells and cancer cells.
Conclusion: Our findings demonstrate that Lrig1 could serve as a potential marker in the prognosis of patients with HCC. We also revealed that Lrig1 might be involved in the metastatic progression of liver cancer. However, its clinical value should be further investigated in the future.
Keywords: Lrig1, hepatocellular carcinoma, prognosis, immunohistochemistry, siRNA, invasion, migration
Introduction
Hepatocellular carcinoma (HCC), a major pathological type of primary liver cancer, is one of the serious threats to human health and the third leading cause of cancer-related deaths worldwide.1,2 The highest incidence rate of HCC has been reported in the developing world and is steadily increasing across the developed world.3 Currently, surgical operation is the most effective treatment of HCC. However, after hepatic resection and liver transplantation, the recurrence incidence and prognosis are still not optimistic. Totally 65%–80% of the cases recur after surgical therapy, and the crucial postoperative 5-year survival rate ranges from 30% to 40%.4,5 The late diagnosis for this rapidly progressing tumor still remains an obstacle in the improvement of prognosis for HCC patients. Therefore, identifying an effective biomarker for early diagnosis and improving treatment strategies are required.
The leucine-rich repeats and immunoglobulin-like domains (Lrig) family comprises three paralogous genes, termed Lrig1, Lrig2, and Lrig3, which are widely expressed in human and mouse tissues. Moreover, Lrig1 is a transmembrane protein that is involved in the regulation of cellular signal transduction, whose transcript is located at chromosome 3p14.3, a region frequently deleted in various types of human cancers.6 In the last decade, Lrig1 has been hypothesized to function as a tumor suppressor in vivo and in vitro. Recently, experimental and clinical data have subsequently supported this hypothesis. Numerous previous reports have shown that Lrig1 is downregulated in several types of tumors, such as cutaneous squamous cell carcinoma, renal cell carcinoma, glioblastoma, and breast cancer.7–10 Recently, researchers revealed that the mechanism of Lrig1 may involve the inhibition of the PI3K/Akt and Ras/Raf/ERK signal transduction pathways.11 However, the expression of Lrig1 and its clinical significance in HCC remain unclear. In addition, Yokdang et al12 found that Lrig1 may represent a barrier to epithelial-to-mesenchymal transition (EMT). Depletion of endogenous Lrig1 in human mammary epithelial cells expands the stem cell population, augments mammosphere formation, and accelerates EMT. Lrig1 is frequently methylated in human colorectal cancer. Lower expression of its mRNA and protein may also contribute to tumor growth.13 These findings provide evidence indicating that Lrig1 functions as a key mediator of metastatic progression. However, very little is known of Lrig1 function in this regard.
In the present study, we aimed to clarify the relationship between the expression of Lrig1 and prognosis of HCC patients and the epigenetic regulation and biological functions of Lrig1 in liver cancer. Here, we investigated the expression of Lrig1 in 133 patients with HCC by immunohistochemistry (IHC) and compared three liver cancer cell lines with the normal liver cell line by reverse transcription polymerase chain reaction (RT-PCR). Furthermore, we estimated the correlation of Lrig1 expression with clinicopathological parameters and prognostic indicators of HCC patients. Finally, we downregulated Lrig1 in normal and cancerous liver cell lines in vitro using interfering RNA (iRNA) technology and evaluated the cell proliferation, migration, and invasion after Lrig1 deficiency. Herein, we expect that our research could provide some clues to understand the function of Lrig1 in liver cancer.
Materials and methods
Patient and tissue specimens
A total of 133 pairs of paraffin-embedded primary HCC tissue specimens and adjacent non-cancerous liver tissue specimens were obtained from patients who had undergone surgery in Liaocheng People’s Hospital from January 2006 to January 2010. The clinical diagnoses were made by a hepatoma specialist and were histologically confirmed by a pathology report. None of the patients had undergone any preoperative treatment before surgery, such as transcatheter arterial chemoembolization or chemotherapy. The study was approved by the Human Ethics Committee of Liaocheng People’s Hospital and Tongji Medical College. All patients provided written informed consent.
IHC and analysis
The paraffin-embedded specimens obtained from all HCC patients were cut into 4 μm sections. The sections were then dewaxed with xylene and rehydrated using a series of graded alcohols after being baked. Then, the endogenous peroxidase activity was blocked and the sections were incubated overnight with the Lrig1 rabbit polyclonal antibody (working dilution 1:100, #ab 197985; Abcam, Cambridge, MA, USA). Subsequently, the sections were incubated with horseradish peroxidase (HRP)-conjugated second antibody. Assessment of the staining was made under a light microscope by two experienced pathologists who did not know the exact condition of the patient. Scoring depended on the staining intensity and proportion as previously described.14 For each tissue core, the intensity of staining was categorized as follows: 0, negative; 1, weak; 2, moderate, and 3, strong. Based on the proportion of staining, the degree was scored on a scale of 0 (<5%, absent), 1 (5%–25%, sporadic), 2 (25%–50%, focal), and 3 (>50%, diffuse). The final score of each staining was obtained by multiplying the two scores. The IHC score ranged from 0 to 9. IHC scores less than 4 points were determined as negative.
Cell culture and transfection
The LO2, SMMC-7721, Hep-G2, and Hep-3B cells were provided by the Department of Liver Surgery of Tongji Hospital. The cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin (complete medium) in a humidified atmosphere containing 5% CO2 at 37°C. For transfection, the cells were plated in 24-well culture plates at a density of 1×105 cells/well. Twenty-four hours after plating, the cells were transfected with three different Lrig1 siRNA fragments and negative control siRNA (siNC), which were designed and synthesized by Ribbio (Guangzhou, People’s Republic of China) for an additional 24 h: Lrig1 siRNA-1 (5′-TCTGCAAATGGCAACCATT-3′), Lrig1 siRNA-2 (5′-ACTCTCTGAGATTGACCCT-3′), and Lrig1 siRNA-3 (5′-CGACATCCATTGCATTTAA-3′). The liver cells were transfected separately with siRNAs and siNC by Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) to knockdown the Lrig1.
Quantitative real-time PCR
The total RNA was isolated from the cells using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instructions. RT-PCR was performed using an SYBR Master Mix kit (Takara, Kusatsu, Japan) on the LightCycler 480 Real-Time PCR System (Roche, Basel, Switzerland). The sequences of primers for the Lrig1 were as follows: forward: 5′-AGCTAACCATCTTATGAGTGCC-3′ and reverse: 5′-CTCAGAAGCAGCAAATTCACA-3′. Each sample was measured in triplicate. A mean value was used to determine mRNA levels using the comparative Ct method, using the formula 2−ΔΔCT, and the mRNA levels were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH; forward: 5′-GACAAAATGGTGAAGGTCGGT-3′ and reverse: 5′-GAGGTCAATGAAGGGGTCG-3′).
Cell growth assay
Cell growth ability was exhibited by the cell counting kit-8 (CCK-8) from Dojindo Laboratories (Kumamoto, Japan). Cells were transplanted into a 96-well plate at a density of 8×102 cells/well and were continuously incubated for 24, 48, 72, 96, and 120 h at 37°C in 5% CO2. After incubation, at designed different times, 10 μL of CCK-8 was added to each well, and following incubation for 1 h, the absorbance at 450 nm was measured to calculate the number of cells. The analysis of each well type was repeated six times independently.
Cell migration and invasion assays
Cell culture was performed in transwell chambers (8-μm pore size; Corning Incorporated, Corning, NY, USA). For the migration assay, 5×104 cells were plated into 200 μL RPMI-1640 medium in the upper chamber and were cultured for 36 h. For the invasion assay, the insert membranes were coated with diluted Matrigel (San Jose, BD Biosciences, San Joes, CA, USA), but were cultured under the same conditions. Finally, the membranes were removed and stained with hematoxylin, and images were captured using an inverted microscope (Olympus BX51; Olympus Corporation, Tokyo, Japan).
Western blotting
The protein levels were quantified by standard Western blotting procedures. Protein extracted from cells was separated on 8% sodium dodecyl sulfate polyacrylamide electrophoresis gels and transferred to nitrocellulose membranes. After being blocked with 5% non-fat milk in Tris-buffered saline for 3 h, the membranes were incubated with indicated primary antibodies (dilution 1:200; Bioss, Beijing, People’s Republic of China) at 4°C overnight, followed by incubation with HRP-conjugated secondary antibody for 3 h. GAPDH (Sungene, Tianjin, People’s Republic of China) was used as a loading control for comparison between samples.
Statistical analysis
All statistical analyses were carried out using the SPSS statistical software package (version 16.0; SPSS Inc., Chicago, IL, USA). Survival curves were calculated by Kaplan–Meier analysis and compared using the log-rank test. Correlations between Lrig1 expression and the clinical variables were analyzed using the Pearson’s chi-squared test. The Student’s t-test was used for other comparisons. All P-values were two-sided, and P-values <0.05 were considered to be statistically significant.
Results
Lrig1 has low expression in human HCC tissue and cell lines
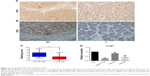
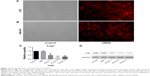
To elucidate the biological significance of Lrig1 in HCC, we examined the expression of Lrig1 in primary paired HCC tissue samples. While normal liver specimens showed positive protein expression, primary liver cancer tissues showed negative protein expression (Figure 1A and B). Low Lrig1 expression was found in 18 of the 133 (13.5%) primary HCC specimens, compared with 68/133 (51.1%) in adjacent non-tumorous (NT) tissues (P<0.05; Figure 1C). In addition, we also compared three HCC cell lines with the normal liver cell line by quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis. Lrig1 expression at the mRNA level in three HCC cell lines evaluated (SMMC-7721, Hep-G2, and Hep-3B) was found to be lower (particularly in SMMC-7721 and Hep-3B cells) compared with the normal cell line LO2 (Figure 1D). Thus, these data suggested that Lrig1 lost was significantly higher in HCC tissues and cell lines than that in NT tissues and cell lines.
Correlation of Lrig1 with clinicopathological variables
To verify the biological function of Lrig1 in HCC, we correlated Lrig1 status in 133 HCC samples with widely recognized clinicopathological features. The low expression of Lrig1 in HCC was associated with tumor size (>5 cm; P<0.01), tumor–node–metastasis (TNM) stage (P<0.05) and tumor recurrence (P<0.01; Table 1). In contrast, Lrig1 expression displayed no association with gender, age, AFP, HBsAg, tumor number, tumor differentiation, and vascular invasion (P>0.05). These data suggest that Lrig1 might play a tumor suppressor role in HCC.
  | Table 1 Correlation of Lrig1 expression with clinicopathological parameters |
Lrig1 is significantly associated with positive prognosis in HCC patients
To examine whether Lrig1 expression is associated with patients’ overall survival, we evaluated via IHC by comparing patients with negative protein expression and those with strong protein expression (n=133). Kaplan–Meier curves showed that HCC patients with high Lrig1 expression had a significantly higher 6-year survival rate than those with negative Lrig1 expression (P<0.05; Figure 2).
Inhibition of Lrig1 expression in HCC cell lines
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis showed relatively higher expression of Lrig1 in Hep-G2 cells than the other two liver cancer cell lines tested (SMMC-7721 and Hep-3B; Figure 1D). Accordingly, we selected LO2 and Hep-G2 as the optimal cells to transfect with three different Lrig1-targeting siRNAs (siLrig1-1, siLrig1-2, and siLrig1-3) and siNC in order to investigate the biological function of Lrig1 in HCC cell lines. The knockdown efficiency of Lrig1 was evaluated by RT-qPCR and Western blot analysis. There is an ~70% decrease in Lrig1 gene expression and Lrig1 expression in siLrig1-2 and siLrig1-3-transfected cells compared with those treated with siLrig1-1 and siNC (Figure 3). Therefore, siLrig1-2 and siLrig1-3 can be chosen in the following studies.
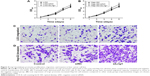
Lrig1 knockdown increases liver cell proliferation, migration, and invasion
Human normal liver cell line (LO2) and HCC cell line (Hep-G2) were treated with siNC and Lrig1-siRNA (siLrig1-2), respectively. As the results in Figure 4 show, the decreased expression of Lrig1 in the Lrig1-siRNA treatment cells significantly promoted the proliferation of the cells, compared with the siNC-treated cells and untreated cells (P<0.05). In migration assay, LO2 cells treated with Lrig1-siRNA showed a high level of penetration through the membranes compared with those treated with siNC and untreated cells (P<0.05). The role of Lrig1 in tumor invasion was apparent in the Lrig1-siRNA treatment Hep-G2 cells, compared with the negative control groups (P<0.01). Taken together, these data suggest that Lrig1 promotes the growth and metastasis of HCC cells in hepatocarcinogenesis. These results further support that Lrig1 is involved in promoting the progression of liver cancer (Figures 3 and S1).
Discussion
In recent years, there are multiple clinical factors considered to be involved in the stratification of HCC patients, including tumor size, serum AFP level, TNM stage, and so on.15 Although these factors can be used to predict prognosis, the prognosis of HCC patients remains dismal.16,17 At present, the diagnosis of HCC is mainly based on biopsy and radiology, but it is unable to discover this disease at the early stage because of the low sensitivity of serum AFP and ultrasonic and other examinations.18 Because of late diagnosis of HCC, only limited therapeutic options with marginal clinical benefits are available for the majority of HCC patients. To further understand the tumorigenesis and development of HCC, novel predictive biomarkers are needed.
Lrig1 is a negative regulator of growth factor signaling that has been confirmed to function as a tumor suppressor in various cancers. Over the years, numerous efforts have been made to look through the relationship between Lrig1 and the pathogenesis and prognosis of tumor patients. In fact, the findings of previous reports have shown that the high expression of Lrig1 is significantly associated with the better prognosis of cancer patients. In a meta-analysis of gene expression and patient outcome in ovarian serous carcinoma, Lrig1 was identified as one single gene prognostic biomarker that predicted survival across all the data sets.19 In an immunohistochemical analysis of 347 non-small cell lung cancer cases, Kvarnbrink et al20 found out that high expression of Lrig1 was an independent prognostic factor that correlated with good survival, particularly among patients with lung adenocarcinoma. However, there are no reports to demonstrate that address the specific function of Lrig1 in the HCC patients. Lrig1 is found low expressed in human HCC tissue and cells in our study. We also analyzed the expression of Lrig1 and its correlation with the clinicopathologic factors and survival rate in 133 total patient-matched tissues. Among the clinicopathological parameters, low Lrig1 expression was significantly correlated with tumor size, TNM stage, and recurrence status. Poor TNM stage and early recurrence status were significantly associated with poor survival of cancer. In brief, negative Lrig1 expression was likely correlated with poor prognosis of liver cancer. To the best of our knowledge, this is the first evidence showing that Lrig1 could be used as a potential positive prognostic marker for HCC patients.
Long-term survival of patients with HCC after curative resection is confounded by its high recurrence rate, which is mainly caused by invasion and metastasis.21 Therefore, identification of metastatic factors and understanding of the underlying molecular pathways involved in metastatic progression are critical. Recently, low expression of Lrig1 was revealed to be an independent risk factor for cancer invasion and metastasis.22–24 The research group of Xie et al25 has found that downregulation of Lrig1 resulted in significantly enhanced capabilities of proliferation and invasion in the glioma cells via activation of epidermal growth factor receptor, protein kinase B (Akt), and c-Myc signaling molecules. It is still unknown whether the Lrig1 expression level plays a role in the regulation of liver cancer cell biological behaviors, such as cell growth and invasion, through some specific signaling pathways. To address these questions, we investigated the biological function of Lrig1 in human liver cancer cells. Downregulation of Lrig1 can promote the growth, invasion and migration of liver cancer cells. These data provided evidence to believe that Lrig1 plays a vital role in cell growth and metastasis and is a promising target for anticancer therapy.
Conclusion
Prior to this study, expression of Lrig1 has never been reported in human liver cancer. Here, our findings of this study also show that Lrig1 may be a potential target in the invasion and metastasis of liver cancer. This suggests a novel approach to investigate the mechanism of the underlying molecular pathways involved in metastatic progression. Furthermore, we demonstrated that low expression of Lrig1 can promote the tumorigenicity of liver cancer. However, its relevance to the prognosis of HCC is poorly understood, and further studies on these mechanisms are still needed in the future.
Acknowledgment
This study was supported by grants from the National Natural Science Foundation of China (Nos 81370581 and 81470936) and the Natural Science Foundation of Hubei Province of China (No 2015CFB573).
Disclosure
The authors report no conflicts of interest in this work.
References
Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27(9):1485–1491. | ||
Zhang HX, Jiang SS, Zhang XF, et al. Protein kinase CK2alpha catalytic subunit is overexpressed and serves as an unfavorable prognostic marker in primary hepatocellular carcinoma. Oncotarget. 2015;6(33):34800–34817. | ||
Thomas MB, Zhu AX. Hepatocellular carcinoma: the need for progress. J Clin Oncol. 2005;23(13):2892–2899. | ||
Kamiyama T, Nakanishi K, Yokoo H, et al. Recurrence patterns after hepatectomy of hepatocellular carcinoma: implication of Milan criteria utilization. Ann Surg Oncol. 2009;16(6):1560–1571. | ||
El-Serag HB, Marrero JA, Rudolph L, Reddy KR. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134(6):1752–1763. | ||
Nilsson J, Vallbo C, Guo D, et al. Cloning, characterization, and expression of human LIG1. Biochem Biophys Res Commun. 2001;284(5):1155–1161. | ||
Thomasson M, Hedman H, Ljungberg B, Henriksson R. Gene expression pattern of the epidermal growth factor receptor family and LRIG1 in renal cell carcinoma. BMC Res Notes. 2012;5:216. | ||
Muller S, Lindquist D, Kanter L, et al. Expression of LRIG1 and LRIG3 correlates with human papillomavirus status and patient survival in cervical adenocarcinoma. Int J Oncol. 2013;42(1):247–252. | ||
Thompson PA, Ljuslinder I, Tsavachidis S, et al. Loss of LRIG1 locus increases risk of early and late relapse of stage I/II breast cancer. Cancer Res. 2014;74(11):2928–2935. | ||
Lindquist D, Kvarnbrink S, Henriksson R, Hedman H. LRIG and cancer prognosis. Acta Oncol. 2014;53(9):1135–1142. | ||
Wang X, He XJ, Xu HQ, Chen ZW, Fan HH. Inhibition of subcutaneously implanted human pituitary tumor cells in nude mice by LRIG1. Genet Mol Res. 2016;15(2). | ||
Yokdang N, Hatakeyama J, Wald JH, et al. LRIG1 opposes epithelial-to-mesenchymal transition and inhibits invasion of basal-like breast cancer cells. Oncogene. 2016;35(22):2932–2947. | ||
Kou C, Zhou T, Han X, Zhuang H, Qian H. LRIG1, a 3p tumor suppressor, represses EGFR signaling and is a novel epigenetic silenced gene in colorectal cancer. Biochem Biophys Res Commun. 2015;464(2):519–525. | ||
Li YS, Chen YZ, Guo XB, Liu X, Li LP. VEZT as a novel independent prognostic factor in gastric cancer. Cancer Biomark. 2015;15(4):375–380. | ||
Toyoda H, Kumada T, Tada T, Sone Y, Kaneoka Y, Maeda A. Tumor markers for hepatocellular carcinoma: simple and significant predictors of outcome in patients with HCC. Liver Cancer. 2015;4(2):126–136. | ||
Chun JM, Kwon HJ, Sohn J, et al. Prognostic factors after early recurrence in patients who underwent curative resection for hepatocellular carcinoma. J Surg Oncol. 2011;103(2):148–151. | ||
Dhanasekaran R, Limaye A, Cabrera R. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis, and therapeutics. Hepat Med. 2012;4:19–37. | ||
Wong GL. Optimal surveillance program for hepatocellular carcinoma – getting ready, but not yet. World J Hepatol. 2015;7(18):2133–2135. | ||
Willis S, Villalobos VM, Gevaert O, et al. Single gene prognostic biomarkers in ovarian cancer: a meta-analysis. PLoS One. 2016;11(2):e0149183. | ||
Kvarnbrink S, Karlsson T, Edlund K, et al. LRIG1 is a prognostic biomarker in non-small cell lung cancer. Acta Oncol. 2015;54(8):1113–1119. | ||
Zhou L, Rui JA, Wang SB, Chen SG, Qu Q. Early recurrence in large hepatocellular carcinoma after curative hepatic resection: prognostic significance and risk factors. Hepatogastroenterology. 2014;61(135):2035–2041. | ||
Mao F, Wang B, Xi G, et al. Effects of RNAi-mediated gene silencing of LRIG1 on proliferation and invasion of glioma cells. J Huazhong Univ Sci Technolog Med Sci. 2012;32(2):227–232. | ||
Mao F, Wang B, Xiao Q, et al. A role for LRIG1 in the regulation of malignant glioma aggressiveness. Int J Oncol. 2013;42(3):1081–1087. | ||
Zhang X, Song Q, Wei C, Qu J. LRIG1 inhibits hypoxia-induced vasculogenic mimicry formation via suppression of the EGFR/PI3K/AKT pathway and epithelial-to-mesenchymal transition in human glioma SHG-44 cells. Cell Stress Chaperones. 2015;20(4):631–641. | ||
Xie R, Yang H, Xiao Q, et al. Downregulation of LRIG1 expression by RNA interference promotes the aggressive properties of glioma cells via EGFR/Akt/c-Myc activation. Oncol Rep. 2013;29(1):177–184. |
Supplementary material
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.