Back to Journals » Infection and Drug Resistance » Volume 12
Loop-mediated isothermal amplification for detection of Legionella pneumophila in respiratory specimens of hospitalized patients in Ahvaz, southwest Iran
Authors Moosavian M , Seyed-Mohammadi S , Saki M , Shahi F , Khoshkholgh Sima M , Afshar D, Barati S
Received 20 December 2018
Accepted for publication 3 February 2019
Published 1 March 2019 Volume 2019:12 Pages 529—534
DOI https://doi.org/10.2147/IDR.S198099
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Mojtaba Moosavian,1,2 Sakineh Seyed-Mohammadi,2,3 Morteza Saki,2,3 Fatemeh Shahi,2,3 Mahtab Khoshkholgh Sima,2 Davoud Afshar,4 Sara Barati5
1Infectious and Tropical Diseases Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 2Department of Microbiology, Faculty of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 3Student Research Committee, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 4Department of Microbiology and Virology, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran; 5Department of Pathobiology, School of Veterinary, University of Shahid Chamran, Ahvaz, Iran
Background: Legionnaires’ disease is an important public health problem that can cause substantial mortality and morbidity. Legionnaires’ disease-risk estimation may be compromised by uncertainties in Legionella-detection methods. The aim of this study was the detection of L. pneumophila in respiratory specimens of hospitalized patients with respiratory symptoms by culture, PCR, and loop-mediated isothermal amplification (LAMP) methods.
Methods: Sputum and bronchoalveolar lavage samples were obtained from patients with pneumonia admitted to teaching hospitals in Ahvaz, Iran from June 2016 to March 2017. Isolation of Legionella spp. was done by culturing the samples directly onto buffered charcoal–yeast extract and modified Wadowsky–Yee agar medium. Then, PCR and LAMP assays were performed for detection of L. pneumophila via its mip gene in respiratory specimens.
Results: A total of 100 respiratory specimens were collected. Our results showed that 1% of the samples were culture positive for Legionella spp., and 3% and 7% of samples were positive for L. pneumophila using the mip gene on PCR and LAMP assays, respectively.
Conclusion: Legionnaires’ disease should be considered in the diagnosis of pulmonary infectious diseases. Also, the LAMP assay is a faster method with higher sensitivity and specificity than conventional methods, such as PCR and culture, for laboratory diagnosis of Legionnaires’ disease.
Keywords: Legionella pneumophila, LAMP, PCR, mip gene
Introduction
Legionnaires’ disease is an important public health problem that causes substantial morbidity and mortality. This bacterial infection is caused primarily by the Gram-negative bacterial genus Legionella.1 Legionella spp. are found in the environment and can enter man-made hot-water systems, air conditioners, and cooling towers of defined facilities, such as hotels, hospitals, and whirlpool spas.2 These bacteria cause community-, travel-, and hospital-acquired pneumonia in humans, usually via inhalation of contaminated aerosols.3 Even with antibiotic therapy, Legionnaires’ disease causes high mortality rates (15%–25%), with elderly and immunocompromised patients being most susceptible.2,4 Approximately 80%–90% of reported cases of Legionnaires’ disease are attributed to L. pneumophila; however, all species may cause infection, especially in immunocompromised hosts.5 Legionnaires’ disease can be potentially fatal if treatment is not undertaken in time, so clinical cases suspected of Legionnaires’ disease should be reported immediately to epidemiological centers to help confirmation of their laboratory diagnoses.6 Over time, the prevalence of Legionnaires’ disease has risen, but the exact incidence of this disease remains unknown, as a result of nonspecific signs and symptoms and variation in diagnostic and surveillance systems among involved countries.1,2
Diagnosis of Legionnaires’ disease is based on culture, serological testing, antigen detection in urine, and nucleic acid–amplification techniques.2 The gold-standard method for identification of Legionella is culture; however, this method has low sensitivity and is time-consuming. Also, direct fluorescent antibody has low sensitivity, and urine-antigen detection is limited to identification of L. pneumophila serogroup 1.7 These limitations have led to the development of rapid molecular methods for detection of Legionella. The current gold standard in molecular diagnosis is based on detection of the mip gene specific for L. pneumophila.8 This gene encodes the peptidylprolyl cis/trans isomerase, and was one of the first genes associated with the ability of Legionella to replicate in eukaryotic cells. mip stands for “macrophage infectivity potentiator”, and mip mutants of L. pneumophila are defective for replication in macrophages, epithelial cells, and amoebae and attenuated in infection.9
Molecular techniques, such as PCR or loop-mediated isothermal amplification (LAMP), can potentially detect Legionella within a few hours in clinical specimens.10 LAMP rapidly amplifies a few DNA molecules with high specificity and efficiency.11 This method has attracted a lot of attention as a potentially simple, accurate, and cost-effective novel nucleic acid–amplification method that has the ability to detect of a wide range of Legionella spp. with high specificity, sensitivity, and rapidity.12,13 The aim of this study was the identification of L. pneumophila from patients with pneumonia symptoms by PCR and LAMP assays as a rapid diagnostic method in comparison with the culture method for laboratory diagnosis of Legionnaires’ disease.
Methods
Ethical considerations
This study was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran (IR.AJUMS.REC.1395.620). Written informed consent was obtained from all patients.
Clinical specimens
Respiratory specimens, including sputum and bronchoalveolar lavage (BAL), were taken by infectious disease specialists from patients with clinical and radiological signs (cough, fever, chill, shortness of breath) suspected of atypical pneumonia who were admitted to teaching hospitals in Ahvaz in the southwest of Iran from June 2016 to March 2017. Samples were transferred to the microbiology laboratory of the School of Medicine in <2 hours in a cold condition.
Isolation of Legionella species
Two respiratory specimens were collected from each patient: one sample inoculated onto the culture media and the other stored at –70°C until molecular assay. Sputum specimens were decontaminated by heat treatment (56°C for 30 minutes), and BAL samples were concentrated by centrifugation. The prepared specimens were cultured onto an unselective medium – buffered charcoal–yeast extract (BCYE) agar (Oxoid, Basingstoke, UK) – and a selective medium, modified Wadowsky–Yee agar (Oxoid) supplemented with L-cysteine. The plates were incubated in candle jars (3%–5% CO2) at 35°C in a humidified atmosphere and inspected for 4–14 days for the presence of Legionella spp. colonies. Colonies showing characteristics of Legionella spp., such as grayish-white, shiny colonies were selected. Gram staining was done to show the thin, faintly stained filamentous Gram-negative morphology. Suspected colonies were subcultured on BCYE agar with and without L-cysteine and unselective media, such as sheep-blood agar and MacConkey agar, for verification. Isolates that grew on BCYE agar with L-cysteine, but not on the other media, were considered Legionella. Strains unable to grow on media without L-cysteine were further identified by PCR and LAMP for the mip gene.14
DNA extraction
Extraction of DNA from cultivated strains, sputum, and BAL samples was performed by High Pure PCR template preparation kit (Roche, Mannheim, Germany) according to the manufacturer’s instructions. The concentration and purity of DNA were determined by absorbance at 260 and 280 nm using NanoDrop spectrophotometry (Thermo Scientific, Waltham, MA, USA) and agarose-gel electrophoresis, respectively.
PCR amplification of mip gene
PCR was employed for the amplification of a 159-bp fragment of the mip gene using specific primers (Table 1).15 The PCR assay was carried out thus: an initial denaturation at 95°C for 4 minutes, followed by 30 cycles at 94°C for 30 seconds, 55°C for 30 seconds, 72°C for 30 seconds, and a final extension at 72°C for 5 minutes. PCR amplification was performed on an Eppendorf thermocycler 5,530 (Roche). In our study, L. pneumophila serogroup 1 (ATCC 33152) was used as the positive control. Amplicons were separated on 1.5% agarose gel prepared in TAE (Tris–acetate–EDTA) buffer and visualized using the gel-documentation system (ProteinSimple, San Jose, CA, USA) after staining with 0.5 µg/mL ethidium bromide (SinaClon, Tehran, Iran).
  | Table 1 Primers used in LAMP and PCR for detection of Legionella pneumophila Abbreviation: LAMP, loop-mediated isothermal amplification. |
Primer design for LAMP assay
To design mip-specific LAMP primers, sequences of the mip gene of Legionella pneumophila subsp. pneumophila Philadelphia 1 strain with gene ID 19832357 was downloaded from the National Center for Biotechnology Information (NCBI) GenBank database. LAMP primers were designed using PrimerExplorer V4 software (http://primerexplorer.jp/e) based on software default settings. Primers included two inner primers, FIP and BIP, and two outer primers, F3 and B3 (Table 1). The specificity of the primers for the mip gene was confirmed using BLAST (basic local alignment search tool) on the NCBI server (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The primers were synthesized commercially by Pishgam (Tehran, Iran).
Optimization of LAMP reaction
Optimization of the LAMP assay was performed in a 25 µL reaction mixture containing 2.5 µL 10× Bst buffer (New England Biolabs, Ipswich, MA, USA), 0.2 pmol each of F3 and B3, 0.8 pmol each of FIP and BIP, different concentrations of dNTPs (0.4, 0.6, 0.8, 1, and 1.2 mM), different concentrations of MgSO4 (3, 4, 5 and 6 mM), 8 U Bst DNA polymerase (New England Biolabs), and different concentrations of DNA template. The reaction mixture was incubated at 60°C–65°C for different times and heated to 95°C for 5 minutes to stop Bst DNA-polymerase activity in the reaction. L. pneumophila serogroup 1 (ATCC 33152) was used as the positive control. Finally, LAMP-reaction products were electrophoresed on 2% agarose gel for detection of best LAMP product. After optimization of the LAMP assay, the amplified products were detected by adding 1 µL 1:10 SYBR green I (Thermo Fisher Scientific) into a reaction microtube. The reaction was considered positive if its color turned from orange to green under ultraviolet light.16
Specificity of LAMP and PCR amplification
The specificity of LAMP and PCR assays was evaluated using different Legionella spp.: L. dumoffii, L. worsleiensis, and L. fairfieldensis. The product was analyzed by gel electrophoresis.
Determination of detection sensitivity in LAMP and PCR assays
For determination of the detection limit, PCR and LAMP reactions were performed using serially diluted DNA templates of L. pneumophila strains, and amplicons were detected by agarose-gel electrophoresis and naked-eye inspection.
Application of LAMP to clinical samples
DNA extracted from 100 clinical respiratory samples was subjected to LAMP and PCR detection for the mip gene. The results of LAMP and PCR assays with the culture were analyzed in comparison.
Statistical analysis
All data were analyzed with SPSS 22. Comparisons between culture, PCR, and LAMP were performed with χ2 using Fisher’s exact test. Independent t-tests were used for analysis of quantitative data between Legionella-positive and -negative groups. Values of P<0.05 were accepted as being statistically significant.
Results
Bacterial isolates by culture method
In this study, 94 sputum samples and 6 BAL samples were collected from patients with pneumonia symptoms. The age of the patients was 15–93 years (average 54 years), including 41 females and 59 males. Results of the culture method showed that one of the sputum samples (1%) was positive for Legionella spp, then confirmed as L. pneumophila by mip-gene PCR. None of the BAL specimens was positive for the presence of Legionella spp. by the culture method.
PCR assay of mip gene
Results of mip PCR revealed that three (3%) sputum samples were positive for 159 bp amplicons. Furthermore, the sample that was positive on culture showed a positive result on PCR. None of the BAL specimens was positive for the presence of mip amplicons on PCR.
Most appropriate factors for LAMP optimization
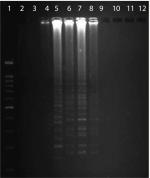
The optimized LAMP assay was carried out thus: a 25 µL reaction mixture containing 2.5 µL 10× Bst buffer, 3 mM MgSO4, 1 µL Bst DNA polymerase (8 IU), 0.8 mM dNTPs, 0.2 pmol each of F3 and B3, 0.8 pmol each of FIP and BIP, 3 µL DNA template, and 9.5 µL distilled deionized water. The best temperature was established as 62°C for 60 minutes. The results of electrophoresis of LAMP-amplification products are shown in Figure 1.
LAMP assay for mip gene
L. pneumophila strains were detected in seven sputum samples (7%) using LAMP (Figure 2). Also, samples that were positive on culture and PCR showed positive results on LAMP assay. L. pneumophila was not found in BAL samples with LAMP. The mean age of patients with positive sputa for L. pneumophila was 57.5 years, with three and four females and males, respectively. These results showed a significant difference in the detection of L. pneumophila by culture and PCR (P=0.031) and also between LAMP and PCR methods (P=0.001). No significant correlation was found among age, sex, and Legionnaires’ disease (P>0.05).
Specificity of LAMP and PCR assays
The specificity of LAMP and PCR assays was tested using DNA samples from different Legionella spp. other than L. pneumophila. The results showed no non-specific amplifications of the mip gene when using DNA from the aforementioned species.
Detection limit of LAMP and conventional PCR
To determine the detection limit of the LAMP and PCR methods, serially diluted DNA templates of L. pneumophila with DNA concentrations ranging from 115 ng/µL to 0.115 pg/µL were provided. SYBR green I (1 μL) was added to the LAMP reaction mixture. Positive reactions would turn green, while negative ones remained orange. Agarose-gel electrophoresis and naked-eye inspection were used to detect the amplified production of PCR and LAMP, respectively. Electrophoresis results showed that the detection limit of PCR for the mip gene was <11.5 pg/µL. The results of naked-eye inspection of LAMP reaction showed that the detection limit of LAMP for mip was <1.15 pg/µL. These results suggested that detection sensitivity of LAMP for mip was tenfold that of PCR.
Discussion
In the present study, three methods were used for detection of Legionella in patients with pneumonia symptoms. We demonstrated that the LAMP assay is a potential diagnostic tool for L. pneumophila detection in sputum specimens. The rate of Legionella spp. detected by culture and PCR methods was 1% and 3%, respectively, while the ability of LAMP was more than the other two methods – 7%. Therefore, LAMP can be useful in specific detection of Legionella.
When comparing PCR to culture, PCR demonstrated more sensitivity (63%). In accordance with our study, Peci et al reported that PCR is a better diagnostic method than urinary antigen and culture for detection of Legionella from BAL, urine, and sputum specimens. They identified L. pneumophila in 3% of BAL fluid and sputum samples by PCR.17
In several studies from Western Europe and North America, 1%–13% of all pneumonia cases were related to Legionella, but because of the difficulty in distinguishing Legionnaires’ disease, many cases probably go unreported.4 Consistently with our results, Cloud et al detected 5.6% Legionella spp. from respiratory specimens by PCR, and stated that the PCR method is a more sensitive and specific method than the culture method for the detection of Legionella in respiratory samples.18 In Weir et al, L. pneumophila was identified in 3.17% of sputum and BAL specimens by PCR. Molecular techniques based on direct extraction and amplification of DNA from respiratory specimens may be useful for the timely diagnosis of Legionnaires’ disease.19
Many studies have shown that the specificity and sensitivity of the LAMP method is greater than that of PCR and culture and other laboratory diagnostic methods.20–22 Initially, we evaluated the specificity of the LAMP assays for these organisms. Four specific primers can recognize the six conserved regions of the target gene, ensuring the high specificity and sensitivity of LAMP. The target DNA fragments can be amplified 109–1010 times within 1 hour under isothermal conditions, indicating that the LAMP assay is stable, rapid, and efficient. Similar to our study, Furuhata et al compared culture and LAMP methods for detection of Legionella spp. in hot-water-bath samples. Legionella spp. were detected in 32% and 60.8% of samples via culture and LAMP, respectively. They indicated that the positive rate in the LAMP test was higher than that in the culture test.20
In a study by Inoue et al, LAMP, PCR, and culture were compared for the rapid detection of Legionella in bathwater samples. A total of 71, 66, and 49 samples were Legionella -positive by the LAMP, PCR, and plate-culture methods, respectively. From these results, it is considered that the LAMP and PCR would be very effective methods for the rapid detection of Legionella.23
Rapid laboratory diagnosis and prompt initiation of appropriate antibiotics for Legionnaires’ disease (community-acquired or hospital-acquired pneumonia) is a crucial approach for management of the disease.24 Therefore, the development of highly sensitive and rapid methods for L. pneumophila detection directly from respiratory samples without the need for sophisticated equipment can improve the management of patients with Legionaries’ disease in regions with limited diagnostic resources. One of these methods is LAMP.7
In comparison of LAMP with PCR and culture, with LAMP as the reference method, LAMP detected four and six L. pneumophila-positive specimens that were not identified by PCR and culture, respectively. This may be attributed to false-negative results of culture and PCR, due to the fastidious nature of Legionella in culture, PCR inhibition, primer mismatch, and the presence of Legionella target below the detection rate.17 LAMP is a powerful amplification assay with a higher detection rate than that of general PCR and other methods.25,26 Furthermore, this cost-effective assay can be performed with limited equipment, even without needing a thermal cycler. According to our results and another study, PCR is a better method than culture, but the LAMP assay is faster and more reliable than both methods for screening of Legionella.
This study had some limitations. The actual prevalence of Legionnaires’ disease cases may be higher than we reported, as we were investigating only L. pneumophila using PCR and LAMP due to its high environmental distribution, while other Legionella spp. can also contribute to Legionnaires’ disease. Additionally, if the urine-antigen method had been used, we probably could have had more ideal comparisons. In this study, due to the fact that the control strains of all Legionella spp were not available, it was not possible to measure the specificity of the LAMP assay with these strains. As such, more Legionella spp. will help for specificity testing of L. pneumophila PCR and LAMP methods. Furthermore, due to a shortage of funding, we were not able to perform DNA sequencing of PCR and LAMP products for validation of molecular identification, in order to avoid false-positive results.
Conclusion
This study showed that Legionnaires’ disease should be considered in the diagnosis of respiratory infectious diseases. We also concluded that the LAMP assay is a faster method with high sensitivity and specificity than conventional methods, such as PCR and culture, for laboratory diagnosis of Legionnaires’ disease. The LAMP assay can be employed in medical centers with limited equipment without the need for a thermocycling apparatus.
Acknowledgments
This study was part of a research project (grant 95135) that was approved and financially supported by the deputy vice-chancellor for research affairs and the Infectious and Tropical Diseases Research Center of Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran, and the authors thank all of them.
Disclosure
The authors report no conflicts of interest in this work.
References
Edelstein PH, Meyer RD. Legionnaires’ disease. a review. Chest. 1984;85(1):114–120. | ||
Tronel H, Hartemann P. Overview of diagnostic and detection methods for legionellosis and Legionella spp. Lett Appl Microbiol. 2009;48(6):653–656. | ||
Rantakokko-Jalava K, Jalava J. Development of conventional and real-time PCR assays for detection of Legionella DNA in respiratory specimens. J Clin Microbiol. 2001;39(8):2904–2910. | ||
Steinert M, Hentschel U, Hacker J. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol Rev. 2002;26(2):149–162. | ||
Yang G, Benson R, Pelish T, Brown E, Winchell JM, Fields B. Dual detection of Legionella pneumophila and Legionella species by real-time PCR targeting the 23S-5S rRNA gene spacer region. Clin Microbiol Infect. 2010;16(3):255–261. | ||
Arora B, Kaur KP, Sethi B. Review article legionellosis: an update. J Clin Diagn Res. 2012;6(7):1331–1336. | ||
Lu X, Mo ZY, Zhao HB, Yan H, Shi L. LAMP-based method for a rapid identification of Legionella spp. and Legionella pneumophila. Appl Microbiol Biotechnol. 2011;92(1):179–187. | ||
Yong SFY, Tan SH, Wee J, et al. Molecular detection of Legionella: moving on from MIP. Front Microbiol. 2010;1:123–127. | ||
Newton HJ, Ang DK, van Driel IR, Hartland EL. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev. 2010;23(2):274–298. | ||
Inoue H, Takama T, Yoshizaki M, Agata K. Detection of Legionella species in environmental water by the quantitative PCR method in combination with ethidium monoazide treatment. Biocontrol Sci. 2015;20(1):71–74. | ||
Karami A, Gill P, Motamedi M, et al. A review of the current isothermal amplification techniques: applications, advantages and disadvantages. J Global Infect Dis. 2011;3(3):293–2. | ||
Annaka T. Rapid and simple detection of Legionella species by LAMP, a new DNA amplification method. J Rapid Methods Autom Microbiol. 2003;14(1):25–30. | ||
Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother. 2009;15(2):62–69. | ||
Tabatabaei M, Hemati Z, Moezzi MO, Azimzadeh N. Isolation and identification of Legionella spp. from different aquatic sources in south-west of Iran by molecular &culture methods. Mol Biol Res Commun. 2016;5(4):215–223. | ||
Wilson DA, Yen-Lieberman B, Reischl U, Gordon SM, Procop GW. Detection of Legionella pneumophila by real-time PCR for the MIP gene. J Clin Microbiol. 2003;41(7):3327–3330. | ||
Trangoni MD, Gioffré AK, Cerón Cucchi ME, et al. LAMP technology: rapid identification of Brucella and Mycobacterium avium subsp. paratuberculosis. Braz J Microbiol. 2015;46(2):619–626. | ||
Peci A, Winter AL, Gubbay JB. Evaluation and comparison of multiple test methods, including real-time PCR, for Legionella detection in clinical specimens. Front Public Health. 2016;4:175–182. | ||
Cloud JL, Carroll KC, Pixton P, Erali M, Hillyard DR. Detection of Legionella species in respiratory specimens using PCR with sequencing confirmation. J Clin Microbiol. 2000;38(5):1709–1712. | ||
Weir SC, Fischer SH, Stock F, Gill VJ. Detection of Legionella by PCR in respiratory specimens using a commercially available kit. Am J Clin Pathol. 1998;110(3):295–300. | ||
Furuhata K, Annaka T, Ikedo M, Fukuyama M, Yoshida S-I. Comparison of loop-mediated isothermal amplification (LAMP) and conventional culture for the detection of Legionella species in hot spring water samples in Japan. Biocontrol Sci. 2005;10(3):117–120. | ||
Seki M, Kilgore PE, Kim EJ, et al. Loop-mediated isothermal amplification methods for diagnosis of bacterial meningitis. Front Pediatr. 2018;6:57–62. | ||
Wang X, Seo DJ, Lee MH, Choi C. Comparison of conventional PCR, multiplex PCR, and loop-mediated isothermal amplification assays for rapid detection of Arcobacter species. J Clin Microbiol. 2014;52(2):557–563. | ||
Inoue H, Noda A, Agata K, et al. Examination of the rapid detection of Legionella from bathwater samples by the loop-mediated isothermal amplification and polymerase chain reaction methods. J Antibacter Antifung Agents. 2004;32. | ||
Phin N, Parry-Ford F, Harrison T, et al. Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect Dis. 2014;14(10):1011–1021. | ||
Lu Q, Zheng W, Luo P, et al. Establishment of loop-mediated isothermal amplification method for detection of Legionella pneumophila. J Zhejiang Univ Sci. 2010;39(3):305–310. | ||
Saharan P, Dhingolia S, Khatri P, et al. Loop-mediated isothermal amplification (LAMP) based detection of bacteria: a review. Afr J Biotechnol. 2014;13(19):1920–1928. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.