Back to Journals » OncoTargets and Therapy » Volume 12
Long noncoding RNA ZEB2-AS1 facilitates laryngeal squamous cell carcinoma progression by miR-6840-3p/PLXNB1 axis
Authors Xu Q, Liu H, Yu B, Chen W, Zhai L, Li X, Fang Y
Received 18 April 2019
Accepted for publication 31 July 2019
Published 6 September 2019 Volume 2019:12 Pages 7337—7345
DOI https://doi.org/10.2147/OTT.S212749
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Qiushi Xu,1 Hongyu Liu,1 Bing Yu,2 Wenjing Chen,2 Lili Zhai,2 XueYing Li,1 Yanchun Fang2
1Ear Nose and Throat Department, Affiliated Qiqihar Hospital, Southern Medical University, The First Hospital of Qiqihaer City, Guangzhou, Heilongjiang 161000, People’s Republic of China; 2Pathology Department, Affiliated Qiqihar Hospital, Southern Medical University, The First Hospital of Qiqihaer City, Guangzhou, Heilongjiang 161000, People’s Republic of China
Correspondence: Yanchun Fang
Pathology Department, Affiliated Qiqihar Hospital, Southern Medical University, The First Hospital of Qiqihaer City, Guangzhou, Heilongjiang 161000, People’s Republic of China
Email [email protected]
Purpose: To investigate the role of zinc finger E‑box‑binding homeobox 2 antisense RNA 1 (ZEB2-AS1) in regulating laryngeal squamous cell carcinoma (LSCC) progression.
Patients and methods: In this retrospective study, we included all patients who underwent a surgical operation at The First Hospital of Qiqihaer City for LSCC. Then, we compared the expression of ZEB2-AS1 in LSCC tissues and paired healthy tissues. Besides, we also performed a series of functional assays, CCK8 assays, colony formation assays, and transwell assays to examine the functions of LSCC cells after knockdown of ZEB2-AS1. Through bioinformatics analysis, we predicted that ZEB2-AS1 binds to miR-6840-3p and targets PLXNB1.
Results: We indicated that the expression of ZEB2-AS1 was higher in LSCC tissues compared to the paired adjacent tissues, and ZEB2-AS1 was also highly expressed in LSCC cell lines. Furthermore, we discovered that ZEB2-AS1 promoted cell proliferation, migration and invasion and was associated with poor prognosis. To find the mechanism, we performed bioinformatics analysis. We identified that ZEB2-AS1 binds to miR-6840-3p and targets PLXNB1. Additionally, miR-6840-3p overexpression or knockdown of PLXNB1 decreased the abilities of cell migration and invasion.
Conclusion: These findings demonstrated that overexpression of ZEB2-AS1 promotes LSCC progression. Overexpression of miR-6840-3p or downregulation of PLXNB1 can abrogate ZEB2-AS1-mediated LSCC malignant development.
Keywords: ZEB2-AS1, LSCC, miR-6840-3p, PLXNB1, migration, invasion
Introduction
Laryngeal squamous cell carcinoma (LSCC) is the most common malignancy with a poor diagnosis.1,2 It has a higher incidence among the head and neck malignancies. In the past years, there are many research focused on the molecular mechanisms about the development and progression of LSCC.3 However, it is still unclear that how LSCC progression. And, it is crucial to identify new therapies for LSCC patients.4
lncRNA was found to have little or no coding potential. However, lncRNA was proven to play an important role in many biological processes including development, immunology, differentiation and cancers.5–8 Moreover, lncRNA was identified to have the potential as a biomarker or therapy target.9 So, it is necessary to explore the role of lncRNA in promoting cancer development.
Zinc finger E‑box‑binding homeobox 2 antisense RNA 1 (ZEB2-AS1) was a noncoding RNA newly found in hepatocellular carcinoma..10 In addition, it has been proved to involve in promoting many cancer progression including lung cancer,11 gastric cancer,12 bladder cancer13 and pancreatic cancer.14 However, the functional role and underlying mechanism in LSCC remain undefined. In our study, we found that ZEB2-AS1 was highly expressed in LSCC tissues and cell lines. And, abnormal expression of ZEB2-AS1 in LSCC cell line promotes the abilities of cell proliferation, migration and invasion.
lncRNA was found to facilitate cancer cell progression through miR-RNA as a ceRNA.6,15 Through bioinformatics analysis, we found that ZEB2-AS1 binds to miR-6840-3p directly. Moreover, we discovered that overexpression of ZEB2-AS1 inhibits miR-6840-3p while inhibition of miR-6840-3p promotes ZEB2-AS1 expression. Moreover, we also discovered that miR-6840-3p targets to PLXNB1.
Plexin-B1 is a transmembrane receptor for semaphoring 4D.16 Previous studies have demonstrated that PLXNB1 is involved in many cellular processes.17,18 And also, there are some reports that proved that PLXNB1 plays a vital role in the progression of glioma.19,20 However, the biological role of PLXNB1 in LSCC is unknown. Through a series of experiments, we revealed that PLXNB1 overexpression associates with LSCC proliferation and invasion.
Materials and methods
Samples
LSCC tissues and paired healthy tissues were isolated from LSCC patients after surgery at The First Hospital of Qiqihaer City. The tissues were frozen in liquid nitrogen. This work was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of The First Hospital of Qiqihaer City. All written informed consents were received from patients.
Cell lines
LSCC cell lines (TU212, Hep-2) and normal bronchial epithelial cell line (16HBE) were obtained from the Chinese Academy of Science of Shanghai (Shanghai, China).
Real-time PCR
RNA was isolated from LSCC tissues and cell lines by Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. qRT-PCR was performed according to the manufacturer’s instructions.
Cell counting kit-8 assays
To measure the cell proliferation abilities, cells were transfected with indicated plasmids and then performed CCK8 assay (7 sea biotech, Shanghai, China).
Colony formation assay
TU212 cells transfected with indicated plasmids were put into 6-well plate. Every well was put into 1000 cells. After 13–15 days culturing, colonies were stained and counted.
Luciferase reporter assay
pMIR-PLXNB1-3ʹUTR (WT or Mutant) plasmids or pMIR-ZEB2-AS1 (WT or Mutant) plasmids and miR-6840-3p mimics were co-transfected in TU212 cells couple with pRL-TK vectors (Promega, USA). After 24 hrs, the dual Glo™ Luciferase Assay System (Promega) was used to measure luciferase activity according to the manufacturer’s protocols.
Western blot
Western blot assay was used to determine the expression of the indicated protein. The assay was performed according to the paper reported.21
Statistical analysis
To analyze the results, we used GraphPad Prism 6 software. The data were shown as means ± SD. Student’s t-test or one-way ANOVA was used to determine significant differences. Survival rate was analyzed by Kaplan–Meier analysis and log-rank test.
Results
ZEB2-AS1 expression is elevated in LSCC tissues and cell lines
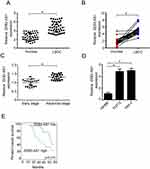
As previous study reports, ZEB2-AS1 promotes many cancer progression. However, the role of ZEB2-AS1 in LSCC is unknown. Firstly, we performed qRT-PCR and found that the expression of ZEB2-AS1 is higher in LSCC samples compared to near healthy tissues (Figure 1A). Among them, 20 (20/27, 74%) pair samples showed a significantly higher expression of ZEB2-AS1 in LSCC tissues compared to adjacent normal tissues (Figure 1B). Next, we examined the expression of ZEB2-AS1 in advanced samples. We found that higher expression associated with advanced cancer samples which indicated that ZEB2-AS1 expression associated with more malignant tumor (Figure 1C). Consistently, we also examined the expression of ZEB2-AS1 in LSCC cell lines and noticed that the expression of ZEB2-AS1 was higher in LSCC cell lines (TU211 and Hep-2) than normal bronchial epithelial cell line (16HBE) (Figure 1D). Then, we wanted to investigate the prognostic significance of ZEB2-AS1. We analyzed the overall survival rate by performing the Kaplan–Meier curve. We divided the 45 patients into higher ZEB2-AS1 expression and lower ZEB2-AS1 expression group based on the median expression level of ZEB2-AS1. Consistently, we found that lower ZEB2-AS1 expression group possessed better overall survival (Figure 1E). Collectively, these data indicate that LSCC samples show a higher ZEB2-AS1 expression which associates with patients’ prognosis.
Knockdown of ZEB2-AS1 significantly decreases LSCC cell migration, invasion and proliferation
To define the role of ZEB2-AS1 in LSCC cells, we performed a series of functional experiments. We constructed shRNAs to decrease the expression of ZEB2-AS1. As examined by qRT-PCR, we found that the expression of ZEB2-AS1 was significantly decreased by the shZEB2-AS1 plasmids (Figure 2A). Then, we found that LSCC cell proliferation ability was obviously decreased in ZEB2-AS1 knockdown group examined by CCK8 assay (Figure 2B). And, we also performed transwell assays. Consistent with the previous study, we found that ZEB2-AS1 knockdown decreased LSCC cell migration and invasion abilities (Figure 2C–D). Colony formation assay was always used to examine the proliferation ability of cancer cells. So, we performed colony formation assay and indicated that LSCC cell proliferation ability significantly decreased after knockdown of ZEB2-AS1 (Figure 2E–F). These data prove that ZEB2-AS1 participates in the regulation of LSCC progression.
Overexpression of ZEB2-AS1 promotes LSCC cells proliferation, migration and invasion
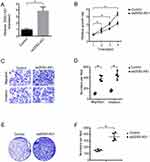
As shown above, we found that knockdown of ZEB2-AS1 decreases LSCC progression. So, we wanted to know the effect of overexpression of ZEB2-AS1. Firstly, we constructed ZEB2-AS1 overexpressed plasmid to elevate the expression of ZEB2-AS1 (Figure 3A). Next, we found that overexpression of ZEB2-AS1 significantly promoted cell proliferation examined by CCK8 assays (Figure 3B). Moreover, we further examined the migration and invasion abilities of LSCC cells. We found that the migration and invasion abilities were increased in ZEB2-AS1 overexpressed LSCC cells (Figure 3C–D). We performed colony formation assay using control and ZEB2-AS1 overexpressed LSCC cells and found that the number of colonies increased significantly in ZEB2-AS1 highly expressed group (Figure 3E–F). Taken together, these data indicate that ZEB2-AS1 significantly promotes the proliferation, migration and invasion abilities of LSCC cells.
ZEB2-AS1 inhibits miR-6840-3p expression to promote LSCC progression through regulating PLXNB1 expression
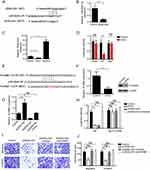
In the previous data, we found that ZEB2-AS1 can promote the proliferation, migration and invasion of LSCC cells. So, we want to explore the mechanism that ZEB2-AS1 regulates LSCC progression. To find the mechanism, we performed bioinformatics analysis. We found that ZEB2-AS1 can form a complementary base pair with miR-6840-3p (Figure 4A). In addition, we found that overexpression of ZEB2-AS1 significantly inhibited miR-6840-3p expression (Figure 4B). In contrast, we also indicated that miR-6840-3p overexpression decreased ZEB2-AS1 expression while miR-6840-3p inhibition promoted ZEB2-AS1 expression (Figure 4C). Next, we performed luciferase assay to prove that ZEB2-AS1 binds to miR-6840-3p. We showed that ectopic expression of miR-6840-3p significantly decreased luciferase intensity in ZEB2-AS1 WT construct while ZEB2-AS1 mutant did not change (Figure 4D).
In addition, we wanted to find the potential target gene regulated by ZEB2-AS1 and miR-6840-3p. We performed bioinformatics analysis again using TargetScan7 program. We showed that miR-6840-3p bound to PLXNB1 through complementary base pair mechanism (Figure 4E). Through qRT-PCR experiment, we found that overexpression of miR-6840-3p decreased PLXNB1 expression. We also proved that by Western blot assay (Figure 4F). Then, we wanted to explore the relationship between ZEB2-AS1, miR-6840-3p and PLXNB1 expression. We indicated that overexpression of ZEB2-AS1 promoted PLXNB1 expression and knockdown of ZEB2-AS1 decreased PLXNB1 expression. Moreover, the increased expression of ZEB2-AS1 can be rescued by ectopic expression of miR-6840-3p (Figure 4G). And, we also performed luciferase assay and found that overexpression of miR-6840-3p or knockdown of ZEB2-AS1 expression decreased luciferase intensity of PLXNB1 WT construct while inhibition of miR-6840-3p coupled with knockdown ZEB2-AS1 expression rescued PLXNB1 WT luciferase intensity. And also, the luciferase intensity of PLXNB1 mutant did not change (Figure 4H). As a result, we also performed transwell assay to examine the migration and invasion abilities. We showed that knockdown of ZEB2-AS1 decreased LSCC cell migration and invasion while inhibition of miR-6840-3p or overexpression of PLXNB1 rescues the abilities of migration and invasion (Figure 4I–J). Collectively, we show that ZEB2-AS1 regulates LSCC progression through binding to miR-6840-3p from PLXNB1 mRNA.
Discussion
In the past decades, lncRNA is proven to play a crucial role in regulating LSCC progression. For example, Yang and colleagues found that lncRNA LOC554202 can promote LSCC carcinoma progression by miR-31.22 They demonstrated that LOC554202 is highly expressed in LSCC tissues while miR-31 is lowly expressed in LSCC tissues compared to adjacent tissues. And also, it is reported that lncRNA UCA1 can activate Wnt/β‑catenin signaling pathway to elevate the proliferation, invasion and migration abilities of LSCC cells.23
ZEB2-AS1 is found to regulate many cancer progression. For instance, ZEB2-AS1 was found to promote breast cancer progression mainly regulating the proliferation and epithelial–mesenchymal transition of breast cancer. And, Wu et al demonstrated that ZEB2-AS1 binds to miR-143-5p and promotes gastric cancer cells proliferation and migration via HIF-1a axis.12 However, the role of ZEB2-AS1 in LSCC remains unclear. So we explore the biological role of ZEB2-AS1 in LSCC progression. In our study, we found that ZEB2-AS1 expresses at a higher level in LSCC tissues compared to near-normal tissues and positively correlated with LSCC progression. Moreover, we found that ZEB2-AS1 regulates the expression of miR-6840-3p and PLXNB1 in LSCC cell line. However, we did not explore the role of ZEB2-AS1/miR-6840-3p/PLXNB1 axis in other cancer cell lines. So we concluded that this mechanism is suited for LSCC. But, to know if this mechanism suit for other cancer needs more evidence.
miRNAs are proven to participant in many cancer regulating. And there are many miRNAs that are involved in regulating LSCC progression.24–26 However, the role of miR-6840-3p has not been reported in any cancer. In our study, we showed that ZEB2-AS1 can bind to miR-6840-3p directly and inhibit the expression of miR-6840-3p. And, we also found that miR-6840-3p mimic can also inhibit ZEB2-AS1 expression. Moreover, miR-6840-3p inhibits the expression of PLXNB1. So, we concluded that ZEB2-AS1 is upstream while miR-6840-3p is downstream.
PLXNB1 is also found to be expressed in many cancer cells. Cao et al found that PLXNB1 can promote the cell proliferation, migration and invasion of cutaneous squamous cell carcinoma.27 In addition, PLXNB1 is proven to regulate Rho/αvβ3/PI3K/Akt signaling pathway, which is involved in regulating glioma invasiveness and angiogenesis.19 In our study, we found that PLXNB1 is a target gene for ZEB2-AS1 and miR-6840-3p. ZEB2-AS1 overexpression or inhibition of miR-6840-3p significantly promotes PLXNB1 expression. And also, overexpression of PLXNB1 restores the decreased migration and invasion abilities of LSCC cells caused by decreased expression of ZEB2-AS1. These data indicated that PLXNB1 plays a crucial role in regulating LSCC progression through ZEB2-AS1 and miR-6840-3p axis.
In conclusion, we showed that ZEB2-AS1 participants in promoting LSCC cells proliferation, migration and invasion via miR-6840-3p/PLXNB1 axis.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Akbaba S, Held T, Lang K, et al. Salvage radiotherapy for recurrent hypopharyngeal and laryngeal squamous cell carcinoma (SCC) after first-line treatment with surgery alone: a 10-year single-centre experience. Radiat Oncol. 2019;14. doi:10.1186/s13014-019-1238-8
2. Boxberg M, Kuhn PH, Reiser M, et al. Tumor budding and cell nest size are highly prognostic in laryngeal and hypopharyngeal squamous cell carcinoma further evidence for a unified histopathologic grading system for squamous cell carcinomas of the upper aerodigestive tract. Am J Surg Pathol. 2019;43:303–313. doi:10.1097/PAS.0000000000001178
3. Malm IJ, Rooper LM, Bishop JA, et al. Molecular and immunologic analysis of laryngeal squamous cell carcinoma in smokers and non-smokers. Am J Otolaryngol. 2019;40:213–217. doi:10.1016/j.amjoto.2018.11.009
4. Cao SJ, Huang YY, Zhang Q, et al. Molecular mechanisms of apoptosis and autophagy elicited by combined treatment with oridonin and cetuximab in laryngeal squamous cell carcinoma. Apoptosis. 2019;24:33–45. doi:10.1007/s10495-018-1497-0
5. Kong XL, Duan Y, Sang YT, et al. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234:9105–9117. doi:10.1002/jcp.27587
6. Liang HL, Zhang CY, Guan HY, Liu J, Cui YB. LncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5p. J Cell Physiol. 2019;234:7266–7278. doi:10.1002/jcp.27484
7. Zhou T, Qin GW, Yang LH, Xiang DK, Li SN. LncRNA XIST regulates myocardial infarction by targeting miR-130a-3p. J Cell Physiol. 2019;234:8659–8667. doi:10.1002/jcp.26327
8. Shi X, Cui ZG, Liu XD, et al. LncRNA FIRRE is activated by MYC and promotes the development of diffuse large B-cell lymphoma via Wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun. 2019;510:594–600. doi:10.1016/j.bbrc.2019.01.105
9. Shen ZS, Li Q, Deng HX, et al. Long non-coding RNA profiling in laryngeal squamous cell carcinoma and its clinical significance: potential biomarkers for LSCC. PLoS One. 2014;9. doi:10.1371/journal.pone.0108237
10. Lan T, Chang L, Wu L, Yuan YF. Downregulation of ZEB2-AS1 decreased tumor growth and metastasis in hepatocellular carcinoma. Mol Med Rep. 2016;14:4606–4612. doi:10.3892/mmr.2016.5836
11. Guo Y, Hu Y, Hu MM, He JB, Li BL. Long non-coding RNA ZEB2-AS1 promotes proliferation and inhibits apoptosis in human lung cancer cells. Oncol Lett. 2018;15:5220–5226. doi:10.3892/ol.2018.7918
12. Wu FX, Gao HY, Liu KG, et al. The IncRNA ZEB2-AS1 is upregulated in gastric cancer and affects cell proliferation and invasion via miR-143-5p/HIF-1 alpha axis. Onco Targets Ther. 2019;12:657–667. doi:10.2147/OTT.S175521
13. Wu XQ, Yan TZ, Wang ZW, Wu X, Cao GH, Zhang C. LncRNA ZEB2-AS1 promotes bladder cancer cell proliferation and inhibits apoptosis by regulating miR-27b. Biomed Pharmacother. 2017;96:299–304. doi:10.1016/j.biopha.2017.08.060
14. Gao H, Gong NN, Ma ZB, et al. LncRNA ZEB2-AS1 promotes pancreatic cancer cell growth and invasion through regulating the miR-204/HMGB1 axis. Int J Biol Macromol. 2018;116:545–551. doi:10.1016/j.ijbiomac.2018.05.044
15. Zhang MB, Li Y, Wang HB, Yu WH, Lin S, Guo JQ. LncRNA SNHG5 affects cell proliferation, metastasis and migration of colorectal cancer through regulating miR-132-3p/CREB5. Cancer Biol Ther. 2019;20:524–536. doi:10.1080/15384047.2018.1537579
16. Malik MFA, Ye L, Jiang WG. Reduced expression of semaphorin 4D and plexin-B in breast cancer is associated with poorer prognosis and the potential linkage with oestrogen receptor. Oncol Rep. 2015;34:1049–1057. doi:10.3892/or.2015.4015
17. Roy AD, Yin T, Choudhary S, Rodionov V, Pilbeam CC, Wu YI. Optogenetic activation of Plexin-B1 reveals contact repulsion between osteoclasts and osteoblasts. Nat Commun. 2017;8:15831.
18. Williamson M, de Winter P, Masters JR. Plexin-B1 signalling promotes androgen receptor translocation to the nucleus. Oncogene. 2016;35:1066–1072. doi:10.1038/onc.2015.160
19. Chang YW, Li L, Zhang LP, et al. Plexin-B1 indirectly affects glioma invasiveness and angiogenesis by regulating the RhoA/alpha v beta 3 signaling pathway and SRPK1. Tumor Biol. 2016;37:11225–11236. doi:10.1007/s13277-016-4849-9
20. Zhang Y, Li Q, Zhuang R, et al. Plexin-B1: a potential diagnostic biomarker for glioma and a future target for glioma immunotherapy. J Neuroimmunol. 2012;252:113–117. doi:10.1016/j.jneuroim.2012.08.005
21. Liu SZ, Duan WC. Long noncoding RNA LINC00339 promotes laryngeal squamous cell carcinoma cell proliferation and invasion via sponging miR-145. J Cell Biochem. 2019;120:8272–8279. doi:10.1002/jcb.v120.5
22. Yang SJ, Wang J, Ge WS, Jiang YF. Long non-coding RNA LOC554202 promotes laryngeal squamous cell carcinoma progression through regulating miR-31. J Cell Biochem. 2018;119:6953–6960. doi:10.1002/jcb.26902
23. Sun SG, Gong C, Yuan K. LncRNA UCA1 promotes cell proliferation, invasion and migration of laryngeal squamous cell carcinoma cells by activating Wnt/-catenin signaling pathway. Exp Ther Med. 2019;17:1182–1189. doi:10.3892/etm.2018.7097
24. Wang Y, Zhang D, Yu CH, et al. MicroRNA-708-5p contributes to the malignant behavior of laryngeal squamous cell carcinoma by directly targeting metastasis suppressor-1. Int J Clin Exp Med. 2019;12:597–603.
25. Zhang F, Cao H. MicroRNA-143-3p suppresses cell growth and invasion in laryngeal squamous cell carcinoma via targeting the k-Ras/Raf/MEK/ERK signaling pathway. Int J Oncol. 2019;54:689–701. doi:10.3892/ijo.2018.4655
26. Han L, Tang MM, Xu XJ, et al. MiR-143-3p suppresses cell proliferation, migration, and invasion by targeting melanoma-associated antigen A9 in laryngeal squamous cell carcinoma. J Cell Biochem. 2019;120:1245–1257. doi:10.1002/jcb.v120.2
27. Cao J, Zhang C, Chen T, et al. Plexin-B1 and semaphorin 4D cooperate to promote cutaneous squamous cell carcinoma cell proliferation, migration and invasion. J Dermatol Sci. 2015;79:127–136. doi:10.1016/j.jdermsci.2015.05.002
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.