Back to Journals » OncoTargets and Therapy » Volume 12
Long Noncoding RNA UCA1 Accelerates Nasopharyngeal Carcinoma Cell Progression By Modulating miR-124-3p/ITGB1 Axis
Received 15 May 2019
Accepted for publication 5 September 2019
Published 11 October 2019 Volume 2019:12 Pages 8455—8466
DOI https://doi.org/10.2147/OTT.S215819
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Takuya Aoki
This paper has been retracted.
Chunxiu Liu,1 Hu Zhang,2 Hui Liu1
1Department of Otolaryngology, Jining First People’s Hospital of Shandong Province, Jinning 272000, People’s Republic of China; 2Department of ENT, Zhangqiu District People’s Hospital, Jinan 250200, People’s Republic of China
Correspondence: Hui Liu
Department of Otolaryngology, Jining First People’s Hospital of Shandong Province, No. 6 Health Road, Rencheng District, Jinning, Shandong 272000, People’s Republic of China
Tel +86-0537-2253830
Email [email protected]
Background: Nasopharyngeal carcinoma (NPC) is a common malignant cancer that is distributed particularly in Southeastern Asia. Previous studies have manifested that long noncoding RNA urothelial carcinoma associated 1 (UCA1) was involved in NPC tumorigenesis and metastasis. However, the biological mechanism of UCA1 for NPC cell progression requires further investigation.
Methods: The expression levels of UCA1, miR-124-3p, integrin beta-1 (ITGB1) were detected by qRT-PCR. Protein expression of ITGB1 was determined by Western blot assay. Cell proliferation, migration and invasion were evaluated by CCK8 and transwell assay, respectively. The interaction between miR-124-3p and UCA1 or ITGB1 was determined by luciferase reporter system, RIP and RNA pull-down assay. Mice model was established by subcutaneously injecting SUNE1 cells stably transfected with sh-UCA1 and sh-NC.
Results: The expression of UCA1 was up-regulated in NPC tissues and cells. However, UCA1 knockdown hindered NPC cell growth, migration and invasion. In addition, the interaction between miR-124-3p and UCA1 or ITGB1 was confirmed by luciferase reporter system, RIP and RNA pull-down assay. Besides, miR-124-3p inhibitor abrogated UCA1 silencing-mediated suppression on cell progression in NPC. Moreover, UCA1 accelerated NPC cell progression through modulating ITGB1 via sponging miR-124-3p. In vivo experiments revealed the interference of UCA1-inhibited tumor growth by regulating miR-124-3p/ITGB1 axis.
Conclusion: UCA1 acts as an oncogene to promote NPC cell proliferation by up-regulating ITGB1 through suppressing miR-124-3p in vitro and in vivo, providing a potential target for NPC diagnosis and treatment.
Keywords: NPC, proliferation, migration, UCA1, miR-124-3p, ITGB1
Introduction
Nasopharyngeal carcinoma (NPC) which originated from nasopharyngeal epithelial cells is one of the most malignant squamous cell carcinomas with high metastasis.1 It geographically distributes in Southeastern Asia and has high incidence in Southern China.2 Clinically, the most prevalent strategies for NPC are still chemotherapy and radiotherapy; however, multidrug resistance and chemotherapy sensitivity could hinder the treatment efficiency and lead to high recurrence, poor therapeutic and prognosis outcomes.3–5 The pathogenesis is complex, including dietary, genetic susceptibility, virus infection and carcinogen hazards.6 Therefore, exploration of the underlying pathological mechanism for NPC cell growth and metastasis is urgently needed.
Long noncoding RNAs (lncRNAs) are RNAs that comprise of over 200 nucleotides in length. Typically, they are involved in multiple biological processes, including cellular signal transmission, chromosome imprinting, hormonal control and genetic translation, therefore imbalance of lncRNAs can trigger different diseases.7,8 Shin Matsubara et al reported that lncRNA-Amhr2 which is located in the cell nucleus is capable of modulating folliculogenesis by activating Amhr2 gene in ovarian granulosa cells.9 In addition, lncRNA-H19 enhanced mesenchymal stem cells angiogenesis and survival by sponging miR-199a-5p.10 Urothelial carcinoma associated 1 (UCA1), derived from bladder cancer, has been identified as oncogenic lncRNA with strong carcinogenic activity.11 Song et al investigated the regulatory mechanism of UCA1 and found UCA1 can positively accelerate colon cancer cells' progression by regulating miR-28-5p/HOXB3 axis.12 However, it remains suspicious whether UCA1 is regulating NPC cell behavior by modulating the specific miRNA.
MicroRNAs (miRNAs) are short-chain noncoding RNAs with 16–22 nucleotides. They participate in various cancer cell functions, for example, cell proliferation, differentiation and apoptosis, by regulating the downstream gene at post-transcriptional level.13 For instance, UCA1 has been reported to promote cell progression via suppressing miR-28-5p and up-regulating HOXB3 expression in colon cancer.14 Binbin Liu et al clarified that miR-124-3p could accelerate intrahepatic cholangiocarcinoma cell progression through regulating UHRF1.15 The role of miR-124-3p in NPC requires in-depth exploration.
Integrin beta-1 (ITGB1), an indispensable member of integrin beta subunit, is involved in the acceleration of cancer cells adhesion, survival and metastasis by interacting with extracellular matrix components fibronectin and laminin.16 Those promotion effects of ITGB1 on cancer cells are regulated by activating intracellular signaling molecules FAK and c-Src to compound pl30Cas and paxillin proteins with kinase activity.17 ITGB1 was verified to stimulate gallbladder cancer (GBC) cells metastasis, while ITGB1 knockdown played an inhibitory role in GNC cell infiltration, proliferation and migration.18 However, the regulatory mechanism of ITGB1 for NPC cell progression is unclear.
In his study, we attempted to illuminate the regulatory effects of UCA1 on NPC tumor growth. The expression of UCA1, miR-124-3p and ITGB1 in NPC was investigated by qRT-PCR. The interaction of miR-124-3p and UCA1 or ITGB1 was validated by dual-luciferase reporter assay. Moreover, animal experiments were conducted to reveal the function of UCA1 in vivo.
Materials And Methods
Tissue Samples
A total of 30 NPC patients were recruited from Jining First People’s Hospital of Shandong Province. NPC patients have not received preoperative therapy. NPC tissues and the adjacent normal tissues were collected from those patients by surgery. All the patients have signed informed consent, and the investigation was approved by the Ethics Committee of Jining First People’s Hospital of Shandong Province, in accordance with the Declaration of Helsinki.
Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
Total RNA extraction was conducted by incubating NPC tissues and cells with Trizol reagent (Invitrogen). RNA reverse transcription reaction was performed using M-MLV reverse transcriptase kit (Invitrogen). qRT-PCR was performed using SYBR green (Applied Biosystems). The primers for miR-124-3p, UCA1 and ITGB1 were listed as follows: UCA1, (Forward, 5ʹ-CTCTCCATTGGGTTCACCATTC-3ʹ; Reverse, 5ʹ-CTCTCCATTGGGTTCACCATTC-3ʹ); miR-124-3p, (Forward, 5ʹ-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGGCATTCA-3ʹ; Reverse, 5ʹ-ACACTCCAGCTGGGTAAGGCACGCGGTGAATGCC-3ʹ); ITGB1 (Forward, 5ʹ-AGGGCCAAATTGTGGGTGG-3ʹ; Reverse, 5ʹ-TGCTGTTCCTTTGCTACGGT-3ʹ).
Cell Transfection
C666-1 cells were purchased from Kalang Biomart, SUNE1 cells and human immortalized nasopharyngeal epithelial cells NP69 were purchased from Sbj-Bio Life Science. SUNE1, C666-1 and NP69 cells were maintained in RPMI-1640 medium (Gibco) supplemented with 10% FBS and 0.05% penicillin/streptomycin. Small interfering RNA (siRNA) targeting UCA1 (si-UCA1, including si-UCA1#1, si-UCA1#2 and si-UCA1#3), siRNA targeting ITGB1 (si-ITGB1), siRNA negative control (si-NC), pcDNA and pcDNA-ITGB1 overexpression vector (ITGB1) were synthesized by Genepharma. The miRNA mimic (miR-124-3p), miR-124-3p inhibitor (in-miR-124-3p), miRNA negative control (miR-NC) and miRNA negative control inhibitor (in-miR-NC) were purchased from RIBOBIO. SUNE1 and C666-1 cells were transfected with those plasmids using Lipofectamine 2000 (Invitrogen).
Cell Viability
After transfected with relative vectors for 24 hrs, SUNE1 and C666-1 cells were seeded in 96-well plates (5000 cells/well) and cultured for another 24 hrs, 48 hrs and 72 hrs. Then, cells were reacted with 10 μL CCK-8 reagent (Beyotime) for 2 hrs. The optical density (OD) value at 450 nm was measured by a microplate reader.
Transwell Assay
Transfected cells were resuspended (2×105 cells/mL, 200 μL) and seeded in the upper chamber pre-treated with Matrigel (Becton Dickinson). The bottom chamber was filled with 600 μL medium containing 10% FBS. After 24 hrs incubation, invasive cells in the lower chamber through the membranes were stained with 0.1% crystal violet (Sigma) for 10 mins and counted using a microscope.
Luciferase Reporter Assay
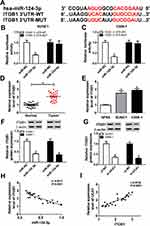
Wild-type and mutant-type luciferase vectors (UCA1 WT, UCA1 MUT, ITGB1 3ʹ-UTR-WT, ITGB1 3ʹ-UTR-MUT) were constructed, then SUNE1 and C666-1 cells were co-transfected with those luciferase vectors and miR-124-3p mimics or miR-NC for 24 hrs using Lipofectamine 2000 transfection reagent. Subsequently, luciferase activities were evaluated by dual-luciferase assay system.
RNA Immunoprecipitation (RIP)
For Ago2 RIP assay, SUNE1 and C666-1 cells were transfected with miR-124-3p or miR-NC and the analysis was performed using EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore). In brief, cells were lysed with RIP buffer 24 hrs post-transfection, centrifuged at 10,000 g for 5 mins, resuspended and incubated with magnetic beads conjugated with Ago2 or IgG antibody. The immunoprecipitated RNA was subjected to qRT-PCR to detect the enrichment of UCA1.
RNA Pull-Down Assay
Biotinylated UCA1 WT (Bio-UCA1 WT), UCA1 MUT (Bio-UCA1 MUT) and negative control (Bio-NC) that purchased from Sangon were transfected into SUNE1 and C666-1 cells. The cells were lysed and collected 24 hrs post-transfection. After incubation with Dynabeads M-280 Streptavidin (Invitrogen) for 10 mins, the bound RNAs were subjected to qRT-PCR for quantification and analysis.
Murine Xenograft Assay
Male Balb/c nude mice were obtained from Vital River Laboratory Animal Technology and all the animal experiments were conducted in accordance with the guidelines provided by the National Animal Care and Ethics Institution. Animal experiments were approved by the animal experimental ethics committee of Jining First People’s Hospital of Shandong Province. Mice were divided into 2 groups (n=6) randomly and subcutaneously injected with SUNE1 cells (5×106 cells, 100 μL) stably transfected with sh-UCA1 or sh-NC. Tumor volumes were recorded every week for 4 weeks and calculated according to the equation: volume (mm3) = length×width×width/2. The mice were sacrificed at 28 days post-treatment, and the tumors were weighed and collected for the subsequent biological analysis.
Statistical Analysis
All the experiments were repeated at least 3 times and the data were analyzed by SPSS software and GraphPad Prism 7. Student’s t-test and one-way ANOVA were used to compare two or more than two groups. The survival rate was analyzed using the Kaplan-Meier method. The correlation between miR-124-3p and UCA1 or ITGB1 was calculated by Pearson’s correlation coefficient. P value less than 0.05 (P<0.05) was considered statistically significant.
Results
UCA1 Was Up-Regulated In NPC Tissues And Cells
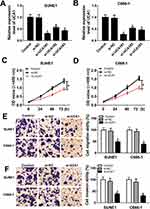
The role of UCA1 in NPC was identified by a series of experiments. As displayed in Table 1, NPC patients at stage III and IV possessed higher UCA1 expression than that of patients at stage I and II. High level of UCA1 was associated with cell metastasis in NPC. UCA1 expression levels in 30 pairs of NPC tissues and corresponding healthy tissues were analyzed by qRT-PCR. The expression of UCA1 was significantly up-regulated in NPC tissues compared with the adjacent normal tissues (P<0.05) (Figure 1A). Similarly, the expression of UCA1 in SUNE1 and C666-1 NPC cells was extremely higher than that in immortalized nasopharyngeal epithelial NP69 cells (Figure 1C). Furthermore, high level of UCA1 contributed to low survival rate, whereas low level of UCA1 showed the opposite result (Figure 1B). Those results collectively manifested that UCA1 was correlated with NPC tumorigenesis.
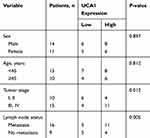 |
Table 1 Analysis Of The Correlation Between Expression Of UCA1 In Primary NPC And Its Clinicopathological Parameters |
UCA1 Knockdown Suppressed NPC Cell Proliferation, Migration And Invasion
To elucidate the biological function of UCA1 in NPC cell progression, SUNE1 and C666-1 cells were transfected with si-NC, si-UCA1#1, si-UCA1#2 and si-UCA1#3. As showed in Figure 2A and B, the expression levels of UCA1 decreased in SUNE1 and C666-1 cells transfected with si-UCA1#1, si-UCA1#2 and si-UCA1#3 compared with their corresponding control groups. Due to the relatively high knockdown efficiency, SUNE1 and C666-1 cells transfected with si-UCA1#1 were recruited for the subsequent cellular experiments. CCK8 results exhibited that UCA1 knockdown slacked NPC cell proliferation ability (Figure 2C and D). In consistent with CCK8 results, transwell assay showed that the migration and invasion ability of SUNE1 and C666-1 cells transfected with si-UCA1 reduced largely (Figure 2E and F). Taken together, UCA1 knockdown attenuated cell proliferation, migration and invasion in NPC.
UCA1 Directly Interacted With miR-124-3p
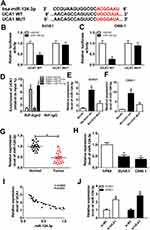
Accumulating evidences have identified lncRNA exerts its regulatory function by interfering with miRNA and further triggering the post-transcriptional process. Bioinformatics analysis prediction by LncBase Predicted v.2 revealed that there were putative binding sites between UCA1 and miR-124-3p (Figure 3A). Dual-luciferase reporter assay in SUNE1 and C666-1 cells revealed that the luciferase activity of cells co-transfected with miR-124-3p mimic and wild-type UCA1 (UCA1-WT) was reduced, whereas the luciferase activity of cells co-transfected with mutant UCA1 (UCA1-MUT) remains unchanged (Figure 3B and C). To further determine the interaction between UCA1 and miR-124-3p, RIP assay was performed. As shown in Figure 3D, the enrichment of UCA1 was enhanced significantly in SUNE1 and C666-1 cells transfected with miR-124-3p in comparison with cells transfected with miR-NC. Similarly, RNA pull-down assay revealed that the expression level of miR-124-3p was up-regulated in SUNE1 and C666-1 cells transfected with UCA1-WT (Figure 3E and F). Then, we discovered that the expression of miR-124-3p was low in NPC tumor tissues and cells compared with the normal tissues and cells (Figure 3G and H), indicating that miR-124-3p functions as a suppressor in NPC cell progression. Subsequently, we found UCA1 was inversely correlated with miR-124-3p (Figure 3I). Furthermore, we detected the expression of miR-124-3p in SUNE1 and C666-1 cells transfected with si-NC and si-UCA1, and observed up-regulation of miR-124-3p level in UCA1 knockdown cells (Figure 3J). Collectively, UCA1 could directly interact with miR-124-3p.
miR-124-3p Inhibitor Abolished UCA1 Silencing-Mediated Regulatory Effects On Cell Proliferation, Migration And Invasion In NPC
Next, to explore the influences of UCA1/miR-124-3p axis on NPC cell proliferation, migration and invasion, SUNE1 and C666-1 cells were transfected with si-NC, si-UCA1, si-UCA1+in-miR-NC and si-UCA1+in-miR-124-3p. Cell viability results showed that the inhibition of miR-124-3p altered UCA1 silencing-mediated inhibitory effects on SUNE1 and C666-1 cell proliferation (Figure 4A and B). In addition, transwell assay exhibited that miR-124-3p inhibitor rescued UCA1 silencing-induced inhibition of cell migration and invasion (Figure 4C and D). Those data demonstrating UCA1 could regulate NPC cell growth by suppressing miR-124-3p expression.
miR-124-3p Exerted Its Function By Targeting ITGB1
Previous studies have identified that miRNA executed their specific regulatory functions through binding to their corresponding target genes. According to bioinformatics analysis, ITGB1 was the potential target gene of miR-124-3p (Figure 5A). Dual-luciferase reporter system analysis further validated the prediction since the luciferase activity reduced enormously in SUNE1 and C666-1 cells co-transfected with miR-124-3p and ITGB1 3ʹ-UTR WT compared with cells co-transfected with miR-NC and ITGB1 3ʹ-UTR MUT (Figure 5B and C). Then, we detected the expression of ITGB1 by qRT-PCR and observed that ITGB1 expression was up-regulated in tumor tissues and NPC cells (SUNE1 and C666-1 cell lines) compared with the adjacent normal tissues and cells (Figure 5D and E). Subsequently, Western blot assay showed that miR-124-3p up-regulation inhibited ITGB1 protein expression whereas UCA1 down-regulation decreased ITGB1 protein expression (Figure 5F and G), revealing that UCA1 regulated ITGB1 by sponging miR-124-3p. Figure 5H and I presented that ITGB1 was positively correlated with UCA1 while inversely correlated with miR-124-3p. Therefore, we considered that ITGB1 was a target of miR-124-3p.
UCA1 Promoted Cell Progression By Up-Regulating ITGB1 Via Sponging miR-124-3p
To verify the regulatory effects of UCA1/miR-124-3p/ITGB1 axis on NPC cell proliferation, migration and invasion, SUNE1 and C666-1 cells were transfected with si-ITGB1+in-miR-124-3p, si-ITGB1+in-miR-NC, si-ITGB1, si-UCA1, si-UCA1+pcDNA, si-UCA1+ITGB1 and si-NC. CCK8 assay and transwell assay indicated that miR-124-3p inhibitor reversed ITGB1 silencing-mediated inhibitory effects on SUNE1 and C666-1 cell proliferation, migration and invasion (Figure 6A–D). Similarly, up-regulation of ITGB1 attenuated UCA1 silencing-induced inhibitory effects on SUNE1 and C666-1 cell progression (Figure 6E–H). Taken together, UCA1 could promote NPC cell development by regulating ITGB1 via sponging miR-124-3p.
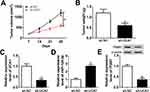
Interference Of UCA1 Suppressed SUNE1 Xenograft Tumor Growth
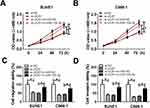
Xenograft model was constructed by subcutaneously injecting SUNE1 cells stably transfected with sh-UCA1 or sh-NC. Tumor growth was suppressed largely in sh-UCA1 xenograft mice compared with sh-NC xenograft mice (Figure 7A). Tumor weight was measured when the mice were sacrificed after 28 days. Reduction of tumor weight was observed in sh-UCA1 xenograft mice (Figure 7B). Furthermore, UCA1 expression was reduced while miR-124-3p expression was enhanced in sh-UCA1 group compared with sh-NC group (Figure 7C and D). Western blot result revealed that ITGB1 protein level was reduced in sh-UCA1 xenograft tissues (Figure 7E). In short, UCA1 knockdown hindered tumor growth in vivo.
Discussion
Recently, lncRNAs have been proposed as promising biomarkers and therapeutic targets of genopathy and cancer due to its variability and abundance.19 Despite lacking protein-encoding capacity, lncRNAs play essential roles in cancer pathogenesis, development and malignancy via participating in gene expression and transcription.20–22 Previous research has confirmed lncRNA LL22NC03-N64E9.1 as lung cancer prognostic biomarker since its sufficiency enhanced lung cancer cells' proliferation.23 Likewise, lncRNA NPCCAT1 overexpression in NPC amplified NPCCAT1 genomic copy number by binding to 3ʹUTR YY1, resulting in accelerating cell adhesion in vivo and in vitro, while NPCCAT1 knockdown reversed the promotion effects.24 However, LncRNA-AF147447 functioned as a suppressor to inhibit gastric cancer formation by binding to MUC2 in gastric cancer.25 UCA1 is located in 19p13.12 chromosome region and involved in cancer deterioration process, for example, UCA1 acts as promoter to induce EMT process and enhance glioma cell invasion by sponging miR-204-5p to up-regulate ZEB1.26 However, the role of UCA1 in NPC cell growth requires further exploration.
In our study, we explored the underlying regulatory mechanism of UCA1 for NPC cell progression. The expression level of UCA1 was elevated in NPC tumor tissue and cell lines measured by qRT-PCR. However, UCA1 silencing led to an obvious suppression on NPC cell proliferation, suggesting that UCA1 regulated NPC cell progression positively. Dual-luciferase reporter assays' results confirmed that UCA1 exerted its function by interacting with miR-124-3p. The interaction between UCA1 and miR-124-3p was further determined by RIP assay and RNA pull-down assay. RIP assay displayed that transfection of miR-124-3p resulted in promoted enrichment of UCA1 in SUNE1 and C666-1 cells compared with transfection of miR-NC. Meanwhile, enhanced miR-124-3p expression was observed in NPC cells transfected with Bio-UCA1 WT in comparison with cells transfected with Bio-UCA1 MUT and Bio-NC. By calculation, we discovered that the UCA1 was correlated with miR-124-3p inversely. Next, rescue experiments revealed that miR-124-3p inhibitors could reverse the inhibitory effects on NPC cells induced by UCA1 silencing. Subsequently, the bioinformatics analysis prediction exhibited that the target gene of miR-124-3p was ITGB1.
Numerous evidences have revealed that ITGB1 acts as an oncogene and participates in the development of many cancers, like lung cancer, hepatocellular carcinoma and glioma.27–29 For instance, Zheng et al elucidated that the up-regulation of ITGB1 in clear cell renal cell carcinoma (ccRCC) has facilitated cell migration via binding the specific site of Mcl-1.30 However, the decreasing of ITGB1 expression inversely reduced trophoblast cell infiltration in preeclampsia patients.31 Therefore, exploration of the potential mechanism of ITGB1 in NPC cell growth and migration possesses great clinical significance.
We further investigated whether miR-124-3p regulates NPC cell development by targeting ITGB1 and altering its post-transcription process. Specifically, overexpression of ITGB1 was observed in NPC tissues and cells, suggesting the oncogenic role of ITGB1. To examine the association between miR-124-3p and ITGB1, Western blot was recruited to detect ITGB1 protein level, resulting in reduced ITGB1 protein expression in SUNE1 and C666-1 cells transfected with miR-124-3p, clarifying that ITGB1 was regulated by miR-124-3p. Similarly, ITGB1 protein expression was down-regulated after UCA1 silencing, implying that UCA1 might function by modulating ITGB1 via miR-124-3p sponging. Moreover, CCK8 assay and wound healing assay revealed that up-regulation of ITGB1attenuated UCA1 silencing-mediated inhibitory effect on NPC cell proliferation, migration and invasion. In vivo experiments further confirmed that UCA1 silencing inhibited NPC cells' progression through regulating miR-124-3p/TGB1 axis. In short, we discovered that UCA1 could facilitate TGB1 expression by sponging miR-124-3p and further accelerate cell progression in NPC.
Conclusion
Our study suggested that UCA1 acted as an oncogene to promote NPC cell progression by up-regulating TGB1 via sponging miR-124-3p. It is the first time to elucidate the biological mechanism of UCA1/miR-124-3p/TGB1 axis for promoting NPC cell growth. Therefore, this study provided promising therapeutical targets for NPC prevention and treatment.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chen J, Li S, Xiao Y, et al. p53R2 as a novel prognostic biomarker in nasopharyngeal carcinoma. BMC Cancer. 2017;17(1):846.
2. Gao J, Shao Z, Yan M, et al. Targeted regulationof STAT3 by miR-29a in mediating Taxol resistance of nasopharyngeal carcinoma cell line CNE-1. Cancer Biomark. 2018;22(4):641–648. doi:10.3233/CBM-170964
3. Liao L, Yan WJ, Tian CM, et al. Knockdown of annexin A1 enhances radioresistance and inhibits apoptosis in nasopharyngeal carcinoma. Technol Cancer Res Treat. 2018;17:1533034617750309. doi:10.1177/1533034617750309
4. Peng H, Zhang J, Zhang PP, et al. ARNTL hypermethylation promotes tumorigenesis and inhibits cisplatin sensitivity by activating CDK5 transcription in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2019;38(1):11. doi:10.1186/s13046-018-0997-7
5. Huang Y, Tian Y, Zhao Y, et al. Efficacy of the hypoxia-activated prodrug evofosfamide (TH-302) in nasopharyngeal carcinoma in vitro and in vivo. Cancer Commun (Lond). 2018;38(1):15. doi:10.1186/s40880-018-0285-0
6. Kong M, Lim YJ, Kim Y. Concurrent chemoradiotherapy for loco-regionally advanced nasopharyngeal carcinoma: treatment outcomes and prognostic factors. Asian Pac J Cancer Prev. 2018;19(6):1591–1599. doi:10.22034/APJCP.2018.19.6.1591
7. Hao S, Yao L, Huang J, et al. Genome-wide analysis identified a number of dysregulated long noncoding RNA (lncRNA) in human pancreatic ductal adenocarcinoma. Technol Cancer Res Treat. 2018;17:1533034617748429. doi:10.1177/1533034617748429
8. Chen T, Xie W, Xie L, et al. Expression of long noncoding RNA lncRNA-n336928 is correlated with tumor stage and grade and overall survival in bladder cancer. Biochem Biophys Res Commun. 2015;468(4):666–670. doi:10.1016/j.bbrc.2015.11.013
9. Kimura AP, Yoneda R, Kurihara M, et al. A long noncoding RNA, lncRNA-Amhr2, plays a role in Amhr2 gene activation in mouse ovarian granulosa cells. Endocrinology. 2017;158(11):4105–4121. doi:10.1210/en.2017-00619
10. Hou J, Wang L, Wu Q, et al. Long noncoding RNA H19 upregulates vascular endothelial growth factor A to enhance mesenchymal stem cells survival and angiogenic capacity by inhibiting miR-199a-5p. Stem Cell Res Ther. 2018;9(1):109. doi:10.1186/s13287-018-0861-x
11. Hu X, Ma R, Fu W, et al. LncRNA UCA1 sponges miR-206 to exacerbate oxidative stress and apoptosis induced by ox-LDL in human macrophages. J Cell Physiol. 2019. doi:10.1002/jcp.28109
12. Wu J, Du M, Zhang Q, et al. Long noncoding RNA UCA1 promotes the proliferation, invasion, and migration of nasopharyngeal carcinoma cells via modulation of miR-145. Onco Targets Ther. 2018;11:7483–7492. doi:10.2147/OTT.S182290
13. Wu W, He K, Guo Q, et al. SSRP1 promotes colorectal cancer progression and is negatively regulated by miR-28-5p. J Cell Mol Med. 2019;23(5):3118–3129. doi:10.1111/jcmm.14134
14. Cui M, Chen M, Shen Z, et al. LncRNA-UCA1 modulates progression of colon cancer through regulating the miR-28-5p/HOXB3 axis. J Cell Biochem. 2019. doi:10.1002/jcb.27630
15. Zhu M, Wei C, Lin J, et al. UHRF1 is regulated by miR‐124‐3p and promotes cell proliferation in intrahepatic cholangiocarcinoma. J Cell Physiol. 2019;234:19875–19885.
16. Huang L, Li X, Gao W. Long non-coding RNA linc-ITGB1 promotes cell proliferation, migration, and invasion in human hepatoma carcinoma by up-regulating ROCK1. Biosci Rep. 2018;38(5):BSR20181289. doi:10.1042/BSR20181289
17. Shang M, Xu X, Zhang M, et al. Long non-coding RNA linc-ITGB1 promotes cell proliferation and migration in human hepatocellular carcinoma cells. Exp Ther Med. 2017;14(5):4687–4692. doi:10.3892/etm.2017.5143
18. Wang L, Zhang Y, Lv W, et al. Long non-coding RNA Linc-ITGB1 knockdown inhibits cell migration and invasion in GBC-SD/M and GBC-SD gallbladder cancer cell lines. Chem Biol Drug Des. 2015;86(5):1064–1071. doi:10.1111/cbdd.12573
19. Li H, Wang Y. Long noncoding RNA (lncRNA) MIR22HG suppresses gastric cancer progression through attenuating NOTCH2 signaling. Med Sci Monit. 2019;25:656–665. doi:10.12659/MSM.912813
20. Xiao B, Huang Z, Zhou R, et al. The prognostic value of expression of the long noncoding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) in patients with solid malignant tumors: a systematic review and meta-analysis. Med Sci Monit. 2018;24:5462–5472. doi:10.12659/MSM.911687
21. M L L, Zhang Q, Yuan X, et al. Long noncoding RNA RP4 functions as a competing endogenous RNA through miR-7-5p sponge activity in colorectal cancer. World J Gastroenterol. 2018;24(9):1004–1012. doi:10.3748/wjg.v24.i9.1004
22. Zhang J, Zhang C. Silence of long non-coding RNA UCA1 inhibits hemangioma cells growth, migration and invasion by up-regulation of miR-200c. Life Sci. 2019;226:33–46.
23. Jing H, Qu X, Liu L, et al. A novel long noncoding RNA (lncRNA), LL22NC03-N64E9.1, promotes the proliferation of lung cancer cells and is a potential prognostic molecular biomarker for lung cancer. Med Sci Monit. 2018;24:4317–4323. doi:10.12659/MSM.908359
24. Su H, Liu L, Zhang Y, et al. Long noncoding RNA NPCCAT1 promotes nasopharyngeal carcinoma progression via upregulating YY1. Biochimie. 2019;157:184–194. doi:10.1016/j.biochi.2018.11.014
25. Zhou X, Chen H, Gu H, et al. Helicobacter pylori infection related long noncoding RNA (lncRNA) AF147447 inhibits gastric cancer proliferation and invasion by targeting MUC2 and up-regulating miR-34c. Oncotarget. 2016;7(50). doi:10.18632/oncotarget.13165
26. Liang C, Yang Y, Guan J, et al. LncRNA UCA1 sponges miR-204-5p to promote migration, invasion and epithelial-mesenchymal transition of glioma cells via upregulation of ZEB1. Pathol Res Pract. 2018;214(9):1474–1481. doi:10.1016/j.prp.2018.07.036
27. Zhu Liang RK, He Z, Lin L-Y, et al. High expression of miR-493-5p positively correlates with clinical prognosis of non small cell lung cancer by targeting oncogene ITGB1. Oncotarget. 2017;8(29):47389–47399. doi:10.18632/oncotarget.17650
28. Zhang L, Zhang T, Deng Z, et al. MicroRNA3653 inhibits the growth and metastasis of hepatocellular carcinoma by inhibiting ITGB1. Oncol Rep. 2019;41(3):1669–1677. doi:10.3892/or.2019.6971
29. Yan M, Zhang L, Li G, et al. Long noncoding RNA linc-ITGB1 promotes cell migration and invasion in human breast cancer. Biotechnol Appl Biochem. 2017;64(1):5–13. doi:10.1002/bab.1461
30. Zheng X-L, Zhang -Y-Y, Lv W-G. Long noncoding RNA ITGB1 promotes migration and invasion of clear cell renal cell carcinoma by downregulating Mcl-1. Eur Rev Med Pharmacol Sci. 2019;23:1996–2002. doi:10.26355/eurrev_201903_17238
31. Zou A-X, Chen B, Li Q-X, Liang Y-C. MiR-134 inhibits infiltration of trophoblast cells in placenta of patients with preeclampsia by decreasing ITGB1 expression. Eur Rev Med Pharmacol Sci. 2018;22:2199–2206. doi:10.26355/eurrev_201804_14804
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.