Back to Journals » OncoTargets and Therapy » Volume 13
Long Noncoding RNA SNHG7, a Molecular Sponge for microRNA-485, Promotes the Aggressive Behavior of Cervical Cancer by Regulating PAK4
Authors Wu F, Sui Y, Wang Y, Xu T, Fan L, Zhu H
Received 26 September 2019
Accepted for publication 22 November 2019
Published 23 January 2020 Volume 2020:13 Pages 685—699
DOI https://doi.org/10.2147/OTT.S232542
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
This paper has been retracted.
Fei Wu, 1 Yujie Sui, 2 Yinhuai Wang, 1 Tianmin Xu, 1 Limei Fan, 1 He Zhu 1
1Department of Gynecology and Obstetrics, The Second Hospital of Jilin University, Changchun 130041, People’s Republic of China; 2Medical Research Center, The Second Hospital of Jilin University, Changchun 130041, People’s Republic of China
Correspondence: He Zhu
Department of Gynecology and Obstetrics, The Second Hospital of Jilin University, 218 Ziqiang Road, Jilin, Changchun 130041, People’s Republic of China
Tel +86 135 1440 0868
Email [email protected]
Purpose: A long noncoding RNA called small nucleolar RNA host gene 7 (SNHG7) is known to be a key regulator of biological processes in multiple human cancer types. In this study, our aims were to determine the expression status of SNHG7 in cervical cancer, to figure out the detailed roles of SNHG7 in cervical cancer cells, and to identify the mechanism underlying the activity of SNHG7 in cervical cancer.
Methods: Reverse-transcription quantitative PCR was performed to measure SNHG7 expression in cervical cancer. A Cell Counting Kit-8 assay, flow-cytometric analysis, cell migration and invasion assays, and a tumor xenograft experiment were conducted to respectively determine the effects of SNHG7 on cervical cancer cell proliferation, apoptosis, migration, and invasion in vitro and tumor growth in vivo.
Results: SNHG7 was found to be markedly upregulated in cervical cancer tissues and cell lines. Higher SNHG7 expression significantly correlated with FIGO stage, lymph node metastasis, the depth of cervical invasion, and shorter overall survival in patients with cervical cancer. Functional experiments indicated that a SNHG7 knockdown attenuated proliferation, migration, and invasiveness and promoted apoptosis of cervical cancer cells in vitro. The SNHG7 knockdown also slowed tumor growth in vivo. Further investigation showed that SNHG7 acts as a competing endogenous RNA for microRNA-485 (miR-485) in cervical cancer cells, and the inhibitory actions of the SNHG7 knockdown on the malignant phenotype were reversed by miR-485 inhibition. P21-activated kinase 4 (PAK4) was identified as a direct target gene of miR-485 in cervical cancer, and PAK4 expression was promoted by SNHG7.
Conclusion: SNHG7 functions as an oncogenic RNA in cervical cancer, competitively binds to miR-485, and thereby upregulates PAK4. This SNHG7–miR-485–PAK4 regulatory network may provide insights into the pathogenesis of cervical cancer, and can help in the identification of novel diagnostic and therapeutic approaches for cervical cancer.
Keywords: small nucleolar RNA host gene 7, P21-activated kinase 4
Introduction
Cervical cancer is the fourth most frequent tumor type and the fourth leading cause of cancer-associated deaths among females worldwide.1 Approximately 530,000 new cervical cancer cases and 250,000 deaths from cervical cancer are reported every year globally, according to World Health Organization (WHO) data.2 At present, the treatment of cervical cancer involves surgical resection and radiation therapy as well as chemotherapy.3 The considerable progress in the diagnosis and treatment has significantly improved the clinical outcomes of patients with cervical cancer; however, many patients receive the diagnosis at advanced stages, and the prognosis of these patients remains unsatisfactory, with a 5-year survival rate of less than 40%.4,5 Furthermore, the molecular pathogenesis of cervical cancer is complicated and needs to be elucidated further.6,7 Therefore, deeper research into the mechanisms underlying the initiation and formation of cervical cancer should help to develop effective therapeutic strategies and improve the clinical outcomes.
Long noncoding RNAs (lncRNAs) are a novel group of single-stranded RNA molecules that are composed of over 200 nucleotides.8 They do not encode proteins, but are widely known to modulate various molecular processes in a tumor.9 Changes in the expression of lncRNAs in cervical cancer have been extensively reported, and have been implicated in carcinogenesis and progression of cervical cancer. For instance, MEG3,10 STXBP5-AS1,11 and LINC0005212 are expressed at low levels in cervical cancer, and suppress malignancy, whereas LUCAT1,13 LINC00511,14 and CASC1115 are upregulated in cervical cancer and play oncogenic roles in the progression of cervical cancer.
MicroRNAs (miRNAs, miRs) are noncoding short RNAs that are approximately 17–22 nucleotides in length.16 MiRNAs regulate gene expression through base-pairing with a complementary site within the 3′-untranslated region (UTR) of target mRNA, thereby causing mRNA degradation and/or translation suppression.17 A variety of studies have revealed that miRNAs are deregulated in cervical cancer, and that their deregulation influences the aggressiveness of cervical cancer.18–20 Accumulating evidence shows that lncRNAs can act as a competitive endogenous RNA (ceRNA) or a molecular sponge for certain miRNAs.21–23 Therefore, the regulatory networks involving lncRNA, miRNA, and mRNA represent promising targets for the diagnosis, prevention, prognosis, and management of cervical cancer.
Recently, lncRNA SNHG7 was reported to be a key regulator of biological processes in multiple human cancer types.24–37 Nonetheless, few studies have addressed the expression and functions of SNHG7 in cervical cancer. Hence, in this study, we attempted to determine the expression status of SNHG7 in cervical cancer, to figure out the detailed roles of SNHG7 in cervical cancer cells, and to identify the mechanism behind the activities of SNHG7 in cervical cancer. We expected to obtain new insights into the SNHG7–miR-485–PAK4 pathway as a novel therapeutic target in human cervical cancer.
Materials and Methods
Tissue Samples from Patients
Cervical cancer tissues and corresponding adjacent normal tissues were collected from 51 patients who underwent surgical resection at The Second Hospital of Jilin University. None of these patients had been treated with preoperative chemoradiotherapy or had a diagnosis of other human cancer types. All the fresh tissue samples were quickly frozen in liquid nitrogen after tissue resection and were stored in liquid nitrogen for further use.
Cell Lines
Four human cervical cancer cell lines (HeLa, SiHa, C-33A, and CaSki) were obtained from the Shanghai Institute of Cell Biology, the Chinese Academy of Sciences (Shanghai, China). Ect1/E6E7, a normal human cervix epithelial cell line, was purchased from the American Type Culture Collection (Manassas, VA, USA). All cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% (v/v) of fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% (v/v) of a solution of antibiotics (final concentration 100 U/mL penicillin and 100 µg/mL streptomycin; Gibco; Thermo Fisher Scientific, Inc.). All the cells were cultured at 37°C in a humidified incubator containing 5% CO2.
A Transfection Procedure
Small interfering RNAs (siRNAs) that were used to silence endogenous SNHG7 expression (si-SNHG7) and negative control (NC) siRNA (si-NC) were purchased from RiboBio Technology (Guangzhou, China). MiR-485 agomir (agomir-485; for increasing miR-485 expression), negative control (NC) agomir (agomir-NC), miR-485 antagomir (antagomir-485; for silencing miR-485 expression), and antagomir-NC were synthesized by GenePharma Co., Ltd. (Shanghai, China). PAK4-overexpressing plasmid pcDNA3.1-PAK4 (hereafter referred to as pc-PAK4) was commercially constructed by the Chinese Academy of Sciences (Changchun, China). Cells were seeded in 6-well plates, and the above-mentioned nucleic acids were introduced into the cells using transfection reagent Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Reverse-Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
The TRIzol Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract total RNA from the tissue samples or cells. To quantify miR-485 expression, reverse transcription was conducted to synthesize cDNA by means of the miScript Reverse Transcription Kit (Qiagen GmbH, Hilden, Germany). Next, this cDNA was subjected to PCR amplification using the miScript SYBR Green PCR Kit (Qiagen GmbH). U6 small nuclear RNA served as an endogenous control of miR-485 expression. To measure PAK4 mRNA and SNHG7 expression, total RNA was reverse-transcribed into cDNA with the PrimeScript RT Reagent Kit (Takara Biotechnology Co., Ltd., Dalian, China). After that, quantitative PCR was carried out via the SYBR Premix Ex Taq™ Kit (Takara Biotechnology Co., Ltd.). All measurements of PAK4 and SNHG7 mRNA levels were conducted using GAPDH as the internal standard. The 2−ΔΔCq method38 was utilized for calculating relative gene expression.
A Cell Counting Kit-8 (CCK-8) Assay
Changes in cell proliferation were determined in the CCK-8 assay. In particular, transfected cells were collected after 24 h cultivation, reseeded in 96-well plates at an initial density of 2 × 103 cells per well, and cultured for 0, 1, 2, or 3 days. At the indicated time points, 10 µL of the CCK-8 solution (Dojindo Laboratories, Kumamoto, Japan) was added into each well, and the cells were incubated at 37°C and 5% CO2 for another 2 h. Finally, absorbance was measured on a spectrophotometer (BioTek, Winooski, VT, USA).
Flow-Cytometric Analysis for Evaluation of Apoptosis
Transfected cells were harvested at 48 h post-transfection through digestion with EDTA-free 0.25% trypsin (Gibco; Thermo Fisher Scientific, Inc.), washed with ice-cold phosphate-buffered saline (Gibco; Thermo Fisher Scientific, Inc.), and subjected to the measurement of apoptosis with the Annexin V-Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit (Biolegend, San Diego, CA, USA). After they were resuspended in 100 µL of binding buffer, cells were double-stained with 5 µL of Annexin V-FITC and 5 µL of a propidium iodide solution, followed by 15 min incubation at room temperature in darkness. The proportion of apoptotic cells was analyzed by means of a FACScan flow cytometer (BD Biosciences, San Jose, CA, USA).
Cell Migration and Invasion Assays
Matrigel (BD Biosciences) diluted in FBS-free DMEM (for coating of the substrate) was incubated with Transwell inserts (Costar, Cambridge, MA, USA), and the resultant inserts were used for a cell invasion assay. Transwell inserts that were not precoated with Matrigel served for the assessment of the cellular migratory ability. For both assays, 5 × 104 transfected cells were resuspended in FBS-free DMEM and then placed in the upper compartments, while DMEM containing 20% of FBS was employed to fill the lower compartments. After 24 h cultivation at 37°C and 5% CO2, the cells remaining on the surface of the upper compartments were wiped off. Prior to staining with 0.1% crystal violet, the inserts were immersed in 4% paraformaldehyde at room temperature for 30 min fixation. Counting of the migratory and invasive cells was carried out under an inverted microscope (IX83; Olympus, Tokyo, Japan). Five visual fields were randomly chosen in every insert, with the average number of migratory and invasive cells calculated accordingly.
Tumor Xenograft Experiments
Female BALB/c nude mice at the age of 4–5 weeks were acquired from the Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). Cells transfected with either si-SNHG7 or si-NC were injected into flank of nude mice via subcutaneous injection. All the mice were maintained under specific pathogen-free conditions and were allowed ad libitum access to water and feed. The dimensions of tumor xenografts were recorded using calipers with an interval of 3 days, after which their volumes were calculated via the standard formula: volume = 0.5 × length × width2. All the mice were euthanized at 4 weeks post-injection, and then, the tumor xenografts were resected for weighing.
Bioinformatic Analysis
Prediction of the interaction between SNHG7 and miRNAs was performed in StarBase 3.0 (http://starbase.sysu.edu.cn/). Three bioinformatic databases, StarBase 3.0, miRanda (http://www.microrna.org) and TargetScan (http://www.targetscan.org), were employed to search for the potential targets of miR-485.
A Luciferase Reporter Assay
Wwild-type (wt) SNHG7 fragments containing the predicted miR-485–binding site and mutant (mut) SNHG7 fragments (ie, with the mutated miR-485–binding site) were designed and synthesized by GenePharma Co., Ltd. The fragments were inserted into the pMIR-REPORT plasmid (Promega, Madison, WI, USA); the resulting luciferase reporter plasmids were named as wt-SNHG7 and mut-SNHG7, respectively. Similar procedures were performed to construct reporter plasmids wt-PAK4 and mut-PAK4. For the reporter assay, either a wt or mut reporter plasmid and either agomir-485 or agomir-NC were cotransfected into cells using transfection reagent Lipofectamine 2000. After 48 h incubation, the transfected cells were assayed using the Dual Luciferase Reporter Assay Kit (Promega) for the evaluation of luciferase activity. The activity of firefly luciferase was normalized to that of Renilla luciferase.
RNA Immunoprecipitation (RIP) Assay
This assay was carried out using the Magna RNA-binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) to evaluate SNHG7–miR-485 interaction in cervical cancer cells. A whole-cell lysate was obtained after lysing the cells with RIP buffer; then, the lysate was incubated with magnetic beads conjugated with a human anti–Argonaute 2 (AGO2) or anti-IgG antibody (Millipore). The coprecipitated RNA was isolated and analyzed by RT-qPCR.
Subcellular Fractionation
Separation of the cytoplasmic and nuclear fractions of cervical cancer cells was conducted with the PARIS Kit (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was extracted from the cytoplasmic and nuclear fractions and was used for determining the SNHG7 distribution.
Western Blot Analysis
Total protein was extracted with the RIPA Protein Extraction Reagent (Beyotime Biotechnology; Shanghai, China), and the protein concentration was quantified by the Bicinchoninic Acid Assay (BCA; Beyotime Biotechnology). Equivalent amounts of protein were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis in a 10% gel and were transferred to a membrane. The latter was blocked at room temperature for 2 h with 5% nonfat dried milk diluted in Tris-buffered saline containing 0.1% of Tween 20 (TBST) and next incubated overnight at 4°C with primary antibodies against PAK4 (sc-390507; 1:1000 dilution; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or GAPDH (sc-47724; 1:1000 dilution; Santa Cruz Biotechnology, Inc.). GAPDH served as the loading control. After extensive washing with TBST, the membranes were probed with a goat anti-mouse IgG antibody conjugated with horseradish peroxidase antibody (sc-516102; 1:5000 dilution; Santa Cruz Biotechnology, Inc.) (secondary antibody) at room temperature for 1 h. The protein bands were visualized by means of the WesternBright Sirius Chemiluminescent Detection Kit (Advansta, California, USA).
Statistical Analysis
All data were presented as means ± standard deviation. The statistical significance of a difference between two groups was examined by Student’s t test. One-way analysis of variance followed by Tukey’s test was carried out for assessing the differences among multiple groups. The correlation between SNHG7 expression and clinical parameters among patients with cervical cancer was assessed via the χ2 test. Kaplan–Meier analysis was conducted to plot survival curves, and the logrank test was performed to evaluate differences between the survival curves. Pairwise expression correlations (SNHG7 vs miR-485 and miR-485 vs PAK4) among cervical cancer tissue samples were determined by Spearman correlation analysis. Data with P < 0.05 were considered statistically significant.
Results
SNHG7 Is Overexpressed in Cervical Cancer and Associated with Poor Clinical Outcomes of Patients with Cervical Cancer
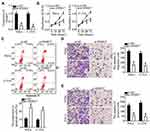
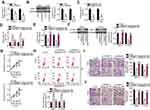
To better understand the functions of SNHG7 in cervical cancer, RT-qPCR was carried out to measure SNHG7 expression in cervical cancer tissue samples and in the corresponding adjacent-normal-tissue samples collected from 51 patients. The results indicated that the expression of SNHG7 was apparently higher in cervical cancer tissue samples than that in the corresponding adjacent normal tissues (Figure 1A, P < 0.05). We then analyzed the clinical value of SNHG7 in patients with cervical cancer. All the 51 patients with cervical cancer were classified into either the high-SNHG7 (n = 26) or low-SNHG7 expression group (n = 25) based on the median value of the SNHG7 levels in the 51 cervical cancer tissue samples. Higher expression of SNHG7 was found to be associated with FIGO stage (P = 0.012), lymph node metastasis (P = 0.001), and the depth of cervical invasion (P = 0.045) in the patients with cervical cancer (Table 1). Survival analysis revealed that the patients with cervical cancer in the high-SNHG7 expression group showed shorter overall survival than did the patients in the low-SNHG7 expression group (Figure 1B, P = 0.024). Furthermore, SNHG7 was overexpressed in all four tested cervical cancer cell lines (HeLa, SiHa, C-33A, and CaSki) relative to that in the normal human cervix epithelial cell line, Ect1/E6E7 (Figure 1C, P < 0.05). HeLa and C-33A cell lines manifested the highest SNHG7 expression among the four tested cervical cancer cell lines; accordingly, these two cell lines were chosen for subsequent experiments. These results meant that SNHG7 was upregulated in cervical cancer tissues and cell lines, implying its high oncogenicity in cervical cancer.
 |
Table 1 Clinicopathological Parameters Associated with Long Noncoding RNA SNHG7 Expression Among Cervical Cancer Patients |
The SNHG7 Knockdown Inhibits the Malignant Characteristics of Cervical Cancer Cells in vitro
To examine the functions of SNHG7 in the malignancy of cervical cancer, the expression of SNHG7 was silenced in HeLa and C-33A cells through si-SNHG7 transfection (Figure 2A, P < 0.05), and si-NC–transfected cells served as the control. The impact of the SNHG7 knockdown on the proliferation of cervical cancer cells was investigated in the CCK-8 assay. The results showed that the knockdown of SNHG7 reduced the capacity for proliferation in HeLa and C-33A cells (Figure 2B, P < 0.05). Flow-cytometric analysis was then conducted to determine the apoptosis status of HeLa and C-33A cells after either si-SNHG7 or si-NC transfection. The proportion of apoptotic HeLa and C-33A cells substantially increased after the SNHG7 knockdown (Figure 2C, P < 0.05). Cell migration and invasion assays were then conducted to evaluate the effects of the SNHG7 knockdown on the migration and invasiveness of cervical cancer cells. The results showed that the reduction in SNHG7 expression noticeably inhibited the migratory (Figure 2D, P < 0.05) and invasive (Figure 2E, P < 0.05) abilities of HeLa and C-33A cells. Consequently, SNHG7 may play an oncogenic part in the progression of cervical cancer in vitro.
SNHG7 Functions as a ceRNA for miR-485 in Cervical Cancer Cells
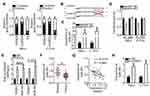
An increasing number of studies indicate that lncRNAs located in the cytoplasm exert their effects on human cancers by acting as a ceRNA for miRNAs.39–41 To illustrate the oncogenic mechanisms of SNHG7 action on cervical cancer cells, we first determined the distribution of SNHG7 within HeLa and C-33A cells. The results suggested that SNHG7 was mainly located in the cytoplasm of HeLa and C-33A cells (Figure 3A), implying that this lncRNA may serve as a ceRNA in cervical cancer cells. Bioinformatic analysis was carried out to search for miRNAs that have complementary sequences for binding to SNHG7. This analysis indicated that miR-485 contains a putative binding site for SNHG7 (Figure 3B). MiR-485 is reported to be downregulated, and to exert tumor-suppressive effects in multiple human cancer types;42–52 consequently, this miRNA was chosen for subsequent experiments. To test whether miR-485 can directly interact with the binding site in SNHG7, we generated the luciferase reporter plasmids harboring either the wild-type or mutant miR-485–binding site. Each of these plasmids was cotransfected with either agomir-485 or agomir-NC into HeLa and C-33A cells. The luciferase reporter assay revealed that the transfection with agomir-485 markedly increased miR-485 expression in HeLa and C-33A cells (Figure 3C, P < 0.05), and that this miRNA significantly decreased the luciferase activity of the wt-SNHG7 plasmid (Figure 3D, P < 0.05). By contrast, the luciferase activity of the mut-SNHG7 plasmid was unaltered in HeLa and C-33A cells after the miR-485 upregulation. Furthermore, the RIP assay was conducted to demonstrate the direct interaction between SNHG7 and miR-485 in cervical cancer cells. MiR-485 and SNHG7 were found to be specifically enriched in an AGO2 immunoprecipitate from the lysates of both HeLa and C-33A cells, as compared with the IgG control group (Figure 3E, P < 0.05). After that, by RT-qPCR, we measured miR-485 expression in the 51 pairs of cervical cancer tissue samples and adjacent-normal-tissue samples. MiR-485 turned out to be downregulated in the cervical cancer tissue samples (Figure 3F, P < 0.05), manifesting an inverse expression correlation with SNHG7 (Figure 3G; r = –0.5579, P < 0.0001). RT-qPCR was finally carried out to quantify miR-485 expression in HeLa and C-33A cells that were transfected with either si-SNHG7 or si-NC. The expression of miR-485 was dramatically higher when SNHG7 was knocked down in HeLa and C-33A cells (Figure 3H, P < 0.05). Thus, SNHG7 may act as a ceRNA for miR-485 in cervical cancer cells.
MiR-485 Directly Targets PAK4 mRNA to Suppress the Aggressive Phenotype of Cervical Cancer Cells
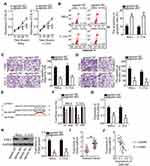
We next investigated the detailed involvement of miR-485 in cervical cancer and elucidated its mechanisms of action. A series of functional experiments was conducted in agomir-485–transfected or agomir-NC–transfected HeLa and C-33A cells and suggested that the ectopic miR-485 expression caused an obvious decrease in cell proliferation (Figure 4A, P < 0.05), an increase in apoptosis (Figure 4B, P < 0.05), and diminution of migration (Figure 4C, P < 0.05) and invasion (Figure 4D, P < 0.05).
Bioinformatics analysis was performed to predict the potential target of miR-485. PAK4 was predicted as a potential target gene of miR-485 (Figure 4E) and was selected for further experiments because PAK4 is closely associated with the malignancy of cervical cancer.53 The luciferase reporter assay next showed that miR-485 overexpression significantly reduced the luciferase activity generated by the wt-PAK4 plasmid (Figure 4F, P < 0.05) but had no influence on the luciferase activity generated by mut-PAK4. In addition, the miR-485 upregulation significantly decreased the expression of PAK4 in HeLa and C-33A cells at both mRNA (Figure 4G, P < 0.05) and protein levels (Figure 4H, P < 0.05). The association between miR-485 and PAK4 was then investigated in the cervical cancer tissue samples. PAK4 mRNA expression was higher in the cervical cancer tissue samples than in the adjacent normal tissues (Figure 4I, P < 0.05). Furthermore, the expression levels of miR-485 and PAK4 mRNA showed a significant negative correlation among the 51 cervical cancer tissue samples, as confirmed by Spearman correlation analysis (Figure 4J; r = –0.6099, P < 0.0001). Collectively, these data proved that miR-485 plays tumor-suppressive roles in cervical cancer cells and that PAK4 is a direct target gene of miR-485 in cervical cancer cells.
Restoration of PAK4 Expression Reverses the Tumor-Suppressive Effects of miR-485 on Cervical Cancer Cells
Rescue experiments were carried out to test whether the decrease in PAK4 expression is essential for the suppressive effects of miR-485 upregulation on the malignant characteristics of cervical cancer cells in vitro. To this end, HeLa and C-33A cells were cotransfected with agomir-485 and either pc-PAK4 or the empty pcDNA3.1 vector. The miR-485 overexpression–mediated decrease in PAK4 mRNA and protein levels was reversed in HeLa and C-33A cells after pc-PAK4 cotransfection (Figure 5A, P < 0.05). Meanwhile, restoration of PAK4 abrogated the tumor-suppressive effects of miR-485 overexpression, such as those involving proliferation (Figure 5B, P < 0.05), apoptosis (Figure 5C, P < 0.05), migration (Figure 5D, P < 0.05), and invasiveness (Figure 5E, P < 0.05) of HeLa and C-33A cells. Collectively, these observations suggested that miR-485 inhibited the malignancy of cervical cancer cells by decreasing PAK4 expression.
SNHG7 Performs Its Oncogenic Functions in Cervical Cancer Cells by Upregulating the Output of the miR-485–PAK4 Axis
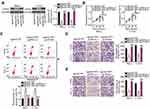
The above results revealed that SNHG7 acts as a ceRNA for miR-485, and that PAK4 is a direct target of miR-485 in cervical cancer cells; accordingly, we next determined whether PAK4 levels can be modulated by SNHG7 in cervical cancer cells through the sponging of miR-485. For this purpose, RT-qPCR and Western blotting were conducted to measure PAK4 mRNA and protein levels in SNHG7-depleted HeLa and C-33A cells, respectively. Knockdown of SNHG7 suppressed the expression of PAK4 mRNA (Figure 6A, P < 0.05) and protein (Figure 6B, P < 0.05) levels in both HeLa and C-33A cells. Next, si-SNHG7 and either antagomir-485 or antagomir-NC were cotransfected into HeLa and C-33A cells, and expression levels of miR-485 and PAK4 protein were determined in cotransfected cells. First, the efficiency of antagomir-485 transfection was evaluated by RT-qPCR (Figure 6C, P < 0.05). The upregulation of miR-485 in HeLa and C-33A cells by the SNHG7 knockdown was attenuated by antagomir-485 cotransfection (Figure 6D, P < 0.05). Similarly, inhibition of miR-485 abrogated the influence of the SNHG7 knockdown on PAK4 mRNA (Figure 6E, P < 0.05) and protein (Figure 6F, P < 0.05) levels in HeLa and C-33A cells. After that, the CCK-8 assay, flow-cytometric analysis, and cell migration and invasion assays were performed, and the results indicated that the SNHG7 knockdown attenuated the proliferation of HeLa and C-33A cells (Figure 6G, P < 0.05), facilitated their apoptosis (Figure 6H, P < 0.05), and restricted their migration (Figure 6I, P < 0.05) and invasion (Figure 6J, P < 0.05), whereas these phenomena were neutralized upon antagomir-485 cotransfection. Altogether, these results suggest that the miR-485–PAK4 axis is necessary for the oncogenic activities of SNHG7 in cervical cancer cells.
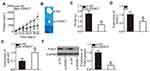
The Knockdown of SNHG7 Inhibits Cervical Cancer Growth in vivo
Tumor xenograft experiments were conducted to assess the participation of SNHG7 in cervical cancer growth in vivo. HeLa cells were transfected with either si-SNHG7 or si-NC and were subcutaneously inoculated into nude mice. Figure 7A indicates that the volume of tumor xenografts in the si-SNHG7 group began to significantly lag behind (as compared with the si-NC group) starting from the 16th day after the inoculation (P < 0.05). Four weeks after the cell injection, all the mice were euthanized, and the tumor xenografts were resected and photographed. Representative images of the tumor xenografts from groups si-SNHG7 and si-NC are presented in Figure 7B. The tumor weight was obviously lower in the si-SNHG7 group than in the si-NC group (Figure 7C, P < 0.05). Furthermore, RT-qPCR analysis of these subcutaneous tumor xenografts revealed that SNHG7 expression was lower (Figure 7D, P < 0.05) while miR-485 expression was higher (Figure 7E, P < 0.05) in the tumor xenografts derived from si-SNHG7–transfected HeLa cells. In addition, we determined the amount of the PAK4 protein in the subcutaneous tumor xenografts from both groups. The results suggested that PAK4 protein expression was notably lower in the si-SNHG7 group than in the si-NC group (Figure 7F, P < 0.05). Collectively, these findings meant that the knockdown of SNHG7 slowed the tumor growth of cervical cancer cells in vivo, and this influence was mediated by downregulation of the miR-485–PAK4 axis output.
Discussion
Nowadays, a variety of lncRNAs are known to be dysregulated in cervical cancer.54–56 Accumulating evidence has confirmed the contribution of lncRNA dysregulation to the aggressiveness of cervical cancer, although only a few lncRNAs have been validated as being involved in cervical cancer.57–59 Therefore, identifying the lncRNAs associated with cervical cancer will improve our knowledge of the role of lncRNAs in the malignancy of cervical cancer, thus highlighting their usefulness as therapeutic and/or prognostic targets in cervical cancer. In this study, we aimed to characterize the expression status, functions, and the mechanisms of action of SNHG7 in cervical cancer. Our study may help identifyan effective, novel, lncRNA-based strategy against cervical cancer.
SNHG7 has been well studied in multiple human cancer types. It is upregulated in breast cancer and positively correlates with tumor stage, lymph node metastasis, and distant metastasis.24 Patients with breast cancer overexpressing SNHG7 show shorter overall survival than do the patients with low SNHG7 expression.24 SNHG7 is overexpressed in prostate cancer tissues and cell lines.25,26 High SNHG7 expression is significantly associated with the Gleason score and tumor stage.25 The expression of SNHG7 is excessive in bladder cancer.27–29 Overexpression of SNHG7 is obviously related to tumor grade, lymph node metastasis, and pathological stage in bladder cancer.27 Furthermore, lung cancer,30,31 gastric cancer,32 glioblastoma,33 colorectal cancer,34,35 esophageal cancer,36 and osteosarcoma37 all overexpress SNHG7. Nevertheless, the expression of SNHG7 in cervical cancer has not been investigated until this study. Here, our major finding is that SNHG7 is upregulated in cervical cancer, and its upregulation notably correlates with FIGO stage, lymph node metastasis, and the depth of cervical invasion. Additionally, patients with cervical cancer in the high-SNHG7 expression group showed shorter overall survival relative to the patients in the low-SNHG7 expression group.
SNHG7 has been confirmed to serve as an oncogenic lncRNA in human cancers. For instance, a knockdown of SNHG7 suppresses the proliferation, migration, and invasiveness of breast cancer cells in vitro and their tumor growth in vivo.24,60,61 An SNHG7 knockdown inhibits prostate cancer cell proliferation, migration, invasion, and epithelial–mesenchymal transition and induces cell cycle arrest at the G0–G1 transition in vitro as well as decreases tumor growth in vivo.25,26 A reduction in SNHG7 expression attenuates bladder cancer cell viability, proliferation, motility, invasion, and epithelial–mesenchymal transition and promotes apoptosis and G0–G1 cell cycle arrest.27–29 Nonetheless, to the best of our knowledge, the functionality of SNHG7 in cervical cancer has not been reported to date. Accordingly, a series of function experiments was conducted here to examine the involvement of SNHG7 in the malignancy of cervical cancer. The results indicated that the knockdown of SNHG7 restricts the proliferation, migration, and invasiveness and promotes the apoptosis of cervical cancer cells. Further investigation revealed that the SNHG7 knockdown retarded tumor cell growth in cervical cancer in vivo.
Recently, lncRNAs have been widely demonstrated to function as ceRNAs, and to directly interact with the mRNA-binding site in miRNAs to attenuate either the translational suppression or degradation of target mRNAs, thereby increasing the mRNA and protein levels of target genes.41,62,63 To investigate whether SNHG7 acts as an miRNA sponge, bioinformatic prediction was performed, and the findings indicated that an miR-485–binding site is present in SNHG7. The luciferase reporter and RIP assays confirmed the direct interaction between SNHG7 and miR-485 in cervical cancer cells. Subsequent experiments revealed that miR-485 is downregulated in cervical cancer tissue samples, and that its expression shows an inverse correlation with that of SNHG7. Furthermore, SNHG7 repressed miR-485 expression, while PAK4 was upregulated by SNHG7 in cervical cancer cells through the sponging of miR-485. Inhibition of miR-485 was found to reverse the inhibitory effect of SNHG7 knockdown with respect to the malignant phenotype of cervical cancer cells. Collectively, these results provide adequate evidence that SNHG7 functions as a ceRNA that sponges miR-485, thereby upregulating PAK4 in cervical cancer.
MiR-485 is underexpressed in a variety of human cancers and exerts tumor-suppressive effects during cancer progression.42–52 Herein, our results confirm that miR-485 is downregulated in both cervical cancer tissues and cell lines. Functionally, miR-485 was found to perform tumor-suppressive activities and was implicated in the control of malignant behavior of cervical cancer cells. After that, we also elucidated the mechanisms underlying the actions of miR-485 in cervical cancer. Our data show that miR-485 directly targets PAK4 mRNA and decreases PAK4 expression in cervical cancer, thereby inhibiting the malignant characteristics. Accordingly, we demonstrated a cervical cancer pathogenesis–related regulatory network, which is composed of SNHG7, miR-485, and PAK4. This information about the SNHG7–miR-485–PAK4 pathway may help to devise promising diagnostic and therapeutic strategies against cervical cancer.
Conclusion
Thus, we identified an oncogenic lncRNA, SNHG7, that plays a role in cervical cancer. SNHG7 increases PAK4 expression in cervical cancer cells by acting as a ceRNA for miR-485, and thus enhances the malignant properties of cervical cancer, thereby hinting at possible SNHG7-based diagnostics and therapeutics.
Abbreviations
3′-UTR, 3′-untranslated region; CCK-8, Cell Counting Kit-8; DMEM, Dulbecco’s Modified Eagle’s Medium; FBS, fetal bovine serum; FITC, fluorescein Isothiocyanate; miRNA, miR, microRNA; mut, mutant; NC, negative control; RT-qPCR, reverse-transcription quantitative PCR; TBST, Tris-buffered saline with 0.1% of Tween 20; wt, wild-type.
Ethics Approval and Informed Consent
This study was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all the participants, and the experiments were conducted with the approval of the Ethics Committee of The Second Hospital of Jilin University (number: 2015.0112). All animal experimental procedures were approved by the Animal Ethics Committee of The Second Hospital of Jilin University (number: 2017.0702), and were performed as per the guidelines of the Animal Protection Law of the People’s Republic of China-2009.
Data Sharing Statement
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Derks M, van der Velden J, de Kroon CD, et al. Surgical treatment of early-stage cervical cancer: a multi-institution experience in 2124 cases in The Netherlands over a 30-year period. Int J Gynecol Cancer. 2018;28(4):757–763. doi:10.1097/IGC.0000000000001228
4. Smith RA, Brooks D, Cokkinides V, Saslow D, Brawley OW. Cancer screening in the United States, 2013: a review of current American Cancer Society guidelines, current issues in cancer screening, and new guidance on cervical cancer screening and lung cancer screening. CA Cancer J Clin. 2013;63(2):88–105. doi:10.3322/caac.21174
5. Kogo R, How C, Chaudary N, et al. The microRNA-218~Survivin axis regulates migration, invasion, and lymph node metastasis in cervical cancer. Oncotarget. 2015;6(2):1090–1100. doi:10.18632/oncotarget.v6i2
6. Bosch FX, de Sanjose S, Chapter 1: human papillomavirus and cervical cancer–burden and assessment of causality. J Natl Cancer Inst Monogr. 2003;31:3–13. doi:10.1093/oxfordjournals.jncimonographs.a003479
7. Yu Y, Zhang Y, Zhang S. MicroRNA-92 regulates cervical tumorigenesis and its expression is upregulated by human papillomavirus-16 E6 in cervical cancer cells. Oncol Lett. 2013;6(2):468–474. doi:10.3892/ol.2013.1404
8. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
9. Schaukowitch K, Kim TK. Emerging epigenetic mechanisms of long non-coding RNAs. Neuroscience. 2014;264:25–38. doi:10.1016/j.neuroscience.2013.12.009
10. Zhang J, Gao Y. Long non-coding RNA MEG3 inhibits cervical cancer cell growth by promoting degradation of P-STAT3 protein via ubiquitination. Cancer Cell Int. 2019;19:175. doi:10.1186/s12935-019-0893-z
11. Shao S, Wang C, Wang S, Zhang H, Zhang Y. LncRNA STXBP5-AS1 suppressed cervical cancer progression via targeting miR-96-5p/PTEN axis. Biomed Pharmacother. 2019;117:109082. doi:10.1016/j.biopha.2019.109082
12. Lin J, Nong LL, Li MQ, Yang FC, Wang SH, Liu MJ. LINC00052 inhibits tumor growth, invasion and metastasis by repressing STAT3 in cervical carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(11):4673–4679. doi:10.26355/eurrev_201906_18047
13. Wang AH, Zhao JM, Du J, Pang QX, Wang MQ. Long noncoding RNA LUCAT1 promotes cervical cancer cell proliferation and invasion by upregulating MTA1. Eur Rev Med Pharmacol Sci. 2019;23(16):6824–6829. doi:10.26355/eurrev_201908_18721
14. Yu CL, Xu XL, Yuan F. LINC00511 is associated with the malignant status and promotes cell proliferation and motility in cervical cancer. Biosci Rep. 2019;39. doi:10.1042/BSR20190903
15. Hsu W, Liu L, Chen X, Zhang Y, Zhu W. LncRNA CASC11 promotes the cervical cancer progression by activating Wnt/beta-catenin signaling pathway. Biol Res. 2019;52(1):33. doi:10.1186/s40659-019-0240-9
16. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi:10.1038/nature09267
17. Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15(12):2070–2079. doi:10.1038/sj.mt.6300311
18. Nahand JS, Taghizadeh-Boroujeni S, Karimzadeh M, et al. microRNAs: new prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J Cell Physiol. 2019;234(10):17064–17099. doi:10.1002/jcp.28457
19. Laengsri V, Kerdpin U, Plabplueng C, Treeratanapiboon L, Nuchnoi P. Cervical cancer markers: epigenetics and microRNAs. Lab Med. 2018;49(2):97–111. doi:10.1093/labmed/lmx080
20. Gonzalez-Quintana V, Palma-Berre L, Campos-Parra AD, et al. MicroRNAs are involved in cervical cancer development, progression, clinical outcome and improvement treatment response (review). Oncol Rep. 2016;35(1):3–12. doi:10.3892/or.2015.4369
21. Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi:10.1186/1476-4598-13-92
22. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310. doi:10.3390/ijms19051310
23. Shuwen H, Qing Z, Yan Z, Xi Y. Competitive endogenous RNA in colorectal cancer: a systematic review. Gene. 2018;645:157–162. doi:10.1016/j.gene.2017.12.036
24. Luo X, Song Y, Tang L, Sun DH, Ji DG. LncRNA SNHG7 promotes development of breast cancer by regulating microRNA-186. Eur Rev Med Pharmacol Sci. 2018;22(22):7788–7797. doi:10.26355/eurrev_201811_16403
25. Qi H, Wen B, Wu Q, et al. Long noncoding RNA SNHG7 accelerates prostate cancer proliferation and cycle progression through cyclin D1 by sponging miR-503. Biomed Pharmacother. 2018;102:326–332. doi:10.1016/j.biopha.2018.03.011
26. Han Y, Hu H, Zhou J. Knockdown of LncRNA SNHG7 inhibited epithelial-mesenchymal transition in prostate cancer though miR-324-3p/WNT2B axis in vitro. Pathol Res Pract. 2019;215:152537. doi:10.1016/j.prp.2019.152537
27. Chen Y, Peng Y, Xu Z, et al. Knockdown of lncRNA SNHG7 inhibited cell proliferation and migration in bladder cancer through activating Wnt/beta-catenin pathway. Pathol Res Pract. 2019;215(2):302–307. doi:10.1016/j.prp.2018.11.015
28. Xu C, Zhou J, Wang Y, et al. Inhibition of malignant human bladder cancer phenotypes through the down-regulation of the long non-coding RNA SNHG7. J Cancer. 2019;10(2):539–546. doi:10.7150/jca.25507
29. Zhong X, Long Z, Wu S, Xiao M, Hu W. LncRNA-SNHG7 regulates proliferation, apoptosis and invasion of bladder cancer cells assurance guidelines. J BUON. 2018;23(3):776–781.
30. She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36(5):2673–2680. doi:10.3892/or.2016.5105
31. Chen K, Abuduwufuer A, Zhang H, Luo L, Suotesiyali M, Zou Y. SNHG7 mediates cisplatin-resistance in non-small cell lung cancer by activating PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2019;23(16):6935–6943. doi:10.26355/eurrev_201908_18733
32. Wang MW, Liu J, Liu Q, et al. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. 2017;21(20):4613–4622.
33. Ren J, Yang Y, Xue J, et al. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun. 2018;496(2):712–718. doi:10.1016/j.bbrc.2018.01.109
34. Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9(7):722. doi:10.1038/s41419-018-0759-7
35. Li Y, Zeng C, Hu J, et al. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J Hematol Oncol. 2018;11(1):89. doi:10.1186/s13045-018-0632-2
36. Xu LJ, Yu XJ, Wei B, et al. LncRNA SNHG7 promotes the proliferation of esophageal cancer cells and inhibits its apoptosis. Eur Rev Med Pharmacol Sci. 2018;22(9):2653–2661. doi:10.26355/eurrev_201805_14961
37. Deng Y, Zhao F, Zhang Z, Sun F, Wang M. Long noncoding RNA SNHG7 promotes the tumor growth and epithelial-to-mesenchymal transition via regulation of miR-34a signals in osteosarcoma. Cancer Biother Radiopharm. 2018;33(9):365–372. doi:10.1089/cbr.2018.2503
38. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
39. Zhao L, Hu K, Cao J, et al. lncRNA miat functions as a ceRNA to upregulate sirt1 by sponging miR-22-3p in HCC cellular senescence. Aging. 2019;11:7098.
40. Long J, Xiong J, Bai Y, et al. Construction and Investigation of a lncRNA-associated ceRNA regulatory network in cholangiocarcinoma. Front Oncol. 2019;9:649. doi:10.3389/fonc.2019.00649
41. Zhou RS, Zhang EX, Sun QF, et al. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer. 2019;19(1):779. doi:10.1186/s12885-019-5983-8
42. Li J, Xu J, Yan X, Jin K, Li W, Zhang R. MicroRNA-485 plays tumour-suppressive roles in colorectal cancer by directly targeting GAB2. Oncol Rep. 2018;40(1):554–564. doi:10.3892/or.2018.6449
43. Hu XX, Xu XN, He BS, et al. microRNA-485-5p functions as a tumor suppressor in colorectal cancer cells by targeting CD147. J Cancer. 2018;9(15):2603–2611. doi:10.7150/jca.24918
44. Jing LL, Mo XM. Reduced miR-485-5p expression predicts poor prognosis in patients with gastric cancer. Eur Rev Med Pharmacol Sci. 2016;20(8):1516–1520.
45. Mao K, Lei D, Zhang H, You C. MicroRNA-485 inhibits malignant biological behaviour of glioblastoma cells by directly targeting PAK4. Int J Oncol. 2017;51(5):1521–1532. doi:10.3892/ijo.2017.4122
46. Wang ZQ, Zhang MY, Deng ML, Weng NQ, Wang HY, Wu SX. Low serum level of miR-485-3p predicts poor survival in patients with glioblastoma. PLoS One. 2017;12(9):e0184969. doi:10.1371/journal.pone.0184969
47. Guo GX, Li QY, Ma WL, Shi ZH, Ren XQ. MicroRNA-485-5p suppresses cell proliferation and invasion in hepatocellular carcinoma by targeting stanniocalcin 2. Int J Clin Exp Pathol. 2015;8(10):12292–12299.
48. Sun X, Liu Y, Li M, Wang M, Wang Y. Involvement of miR-485-5p in hepatocellular carcinoma progression targeting EMMPRIN. Biomed Pharmacother. 2015;72:58–65. doi:10.1016/j.biopha.2015.04.008
49. Mou X, Liu S. MiR-485 inhibits metastasis and EMT of lung adenocarcinoma by targeting Flot2. Biochem Biophys Res Commun. 2016;477(4):521–526. doi:10.1016/j.bbrc.2016.04.043
50. Lou C, Xiao M, Cheng S, et al. MiR-485-3p and miR-485-5p suppress breast cancer cell metastasis by inhibiting PGC-1alpha expression. Cell Death Dis. 2016;7:e2159. doi:10.1038/cddis.2016.27
51. Anaya-Ruiz M, Bandala C, Perez-Santos JL. miR-485 acts as a tumor suppressor by inhibiting cell growth and migration in breast carcinoma T47D cells. Asian Pac J Cancer Prev. 2013;14(6):3757–3760. doi:10.7314/APJCP.2013.14.6.3757
52. Chen Z, Li Q, Wang S, Zhang J. miR4855p inhibits bladder cancer metastasis by targeting HMGA2. Int J Mol Med. 2015;36(4):1136–1142. doi:10.3892/ijmm.2015.2302
53. Shu XR, Wu J, Sun H, Chi LQ, Wang JH. PAK4 confers the malignance of cervical cancers and contributes to the cisplatin-resistance in cervical cancer cells via PI3K/AKT pathway. Diagn Pathol. 2015;10:177. doi:10.1186/s13000-015-0404-z
54. Mao BD, Xu P, Xu P, Zhong Y, Ding WW, Meng QZ. LINC00511 knockdown prevents cervical cancer cell proliferation and reduces resistance to paclitaxel. J Biosci. 2019;44(2). doi:10.1007/s12038-019-9851-0
55. Zhao H, Zheng GH, Li GC, et al. Long noncoding RNA LINC00958 regulates cell sensitivity to radiotherapy through RRM2 by binding to microRNA-5095 in cervical cancer. J Cell Physiol. 2019;234(12):23349–23359. doi:10.1002/jcp.28902
56. Meng Q, Zhang R, Ding W, Mao B. Long noncoding RNA ZFAS1 promotes cell proliferation and tumor growth by upregulating LIN28 in cervical carcinoma. Minerva Med. 2019.
57. Wang Q, Ding J, Nan G, Lyu Y, Ni G. LncRNA NOC2L-4.1 functions as a tumor oncogene in cervical cancer progression by regulating the miR-630/YAP1 pathway. J Cell Biochem. 2019;120(10):16913–16920. doi:10.1002/jcb.28949
58. Fan MJ, Zou YH, He PJ, Zhang S, Sun XM, Li CZ. Long non-coding RNA SPRY4-IT1 promotes epithelial-mesenchymal transition of cervical cancer by regulating the miR-101-3p/ZEB1 axis. Biosci Rep. 2019;39(6). doi:10.1042/BSR20181339
59. Zhu Y, Liu B, Zhang P, Zhang J, Wang L. LncRNA TUSC8 inhibits the invasion and migration of cervical cancer cells via miR-641/PTEN axis. Cell Biol Int. 2019;43(7):781–788. doi:10.1002/cbin.v43.7
60. Sun X, Huang T, Liu Z, Sun M, Luo S. LncRNA SNHG7 contributes to tumorigenesis and progression in breast cancer by interacting with miR-34a through EMT initiation and the Notch-1 pathway. Eur J Pharmacol. 2019;856:172407. doi:10.1016/j.ejphar.2019.172407
61. Gao YT, Zhou YC. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 7 (SNHG7) promotes breast cancer progression by sponging miRNA-381. Eur Rev Med Pharmacol Sci. 2019;23(15):6588–6595. doi:10.26355/eurrev_201908_18545
62. Guo Z, He C, Yang F, Qin L, Lu X, Wu J. Long non-coding RNA-NEAT1, a sponge for miR-98-5p, promotes expression of oncogene HMGA2 in prostate cancer. Biosci Rep. 2019;39. doi:10.1042/BSR20190635
63. Huang QR, Pan XB. Prognostic lncRNAs, miRNAs, and mrnas form a competing endogenous RNA network in colon cancer. Front Oncol. 2019;9:712. doi:10.3389/fonc.2019.00712
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.