Back to Journals » OncoTargets and Therapy » Volume 13
Long Noncoding RNA SNHG6 Functions as an Oncogene in Non-Small Cell Lung Cancer via Modulating ETS1 Signaling
Received 20 October 2019
Accepted for publication 7 December 2019
Published 30 January 2020 Volume 2020:13 Pages 921—930
DOI https://doi.org/10.2147/OTT.S235336
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Hua Geng, Shixiong Li, Meilin Xu
Department of Pathology, Tianjin Chest Hospital, Tianjin 300222, People’s Republic of China
Correspondence: Meilin Xu
Department of Pathology, Tianjin Chest Hospital, No. 261 Taierzhuang South Road, Tianjin 300222, People’s Republic of China
Tel +86 22 8818 5111
Email [email protected]
Background: Non-small cell lung cancer (NSCLC) is a great threat to human health and the biology of the NSCLC still remains largely unknown. Aberrantly expressed long non-coding RNA (lncRNA) Small nucleolar RNA host gene 6 (SNHG6) was involved in the tumorigenesis and progression of various cancers. The aim of this study is to investigate the roles of SNHG6 in NSCLC.
Methods: qRT-PCR and Western blot assays were applied to detect gene expressions. Cell proliferation and migration assays were used to analyze the gene functions. Luciferase reporter assay, RNA Immunoprecipitation assay and Chromatin immunoprecipitation assay were performed to investigate the molecular mechanism.
Results: We found that SNHG6 expression was significantly increased in NSCLC tissues and cell lines and its high expression was correlated with malignant features of NSCLC. In in vitro assays, knockdown of SNHG6 significantly depressed the proliferation vitality and migration activity of NSCLC cells. Research on mechanisms revealed that SNHG6 exerted its tumorigenesis role by promoting ETS1 expression via competitively binding with miR-944 and miR-181d-5p. We also demonstrated that ETS1 enhanced the expression of WIPF1 via binding to its promoter and SNHG6 could thereby regulate the expression of ETS1 target genes including WIPF1, MMP2 and MMP9.
Conclusion: Our study illustrates that SNHG6 is an oncogene in NSCLC and involved in NSCLC tumorigenesis by regulating ETS1 signaling via miR-944 and miR-181d-5p.
Keywords: non-small cell lung cancer, lncRNA, SNHG6, miRNAs, ETS1
Introduction
Lung cancer is the most common malignant tumors worldwide and also the leading cause of cancer deaths worldwide.1,2 Non-small cell lung cancer (NSCLC) is the main type of lung cancer which accounts for approximately 80% of all lung cancer cases. Most NSCLC patients are diagnosed at an advanced stage which causes the prognosis of NSCLC very poor.3,4 Therefore, it is of great importance to further understand the molecular mechanism involved in the occurrence, development and progression of NSCLC.5
Long non-coding RNA (lncRNA) is a class of RNA with over 200 nt in length and no protein-coding function.6 LncRNAs were found to be dysregulated in many cancers and participate in the tumor progressions.7 Recently, more and more studies focused on lncRNAs and investigated their functions in NSCLC.8–10 For example, lncRNA PCAT6 was proved to function as an oncogene by binding to EZH2 and suppressing LATS2 in NSCLC;11 lncRNA AFAP1-AS1 was found to serve as a candidate prognostic biomarker and regulates NSCLC cell proliferation by epigenetically repressing p21 expression.12
Recent researches found that Small nucleolar RNA host gene 6 (SNHG6) could serve as an oncogene in various human cancers, including colorectal cancer,13 ovarian clear cell carcinoma,14 osteosarcoma,15 breast cancer,16 and gastric cancer.17 However, there are still no studies of SNHG6 in NSCLC. So, in the present study, we observed the expression and function of SNHG6 in NSCLC, and investigated the potential molecular mechanism of SNHG6 in NSCLC cell lines.
Materials and Methods
Human Samples
The tumor tissues and adjacent normal tissues (ANTs) were collected from 60 NSCLC patients who accepted radical surgery therapy in the Tianjin Chest Hospital. All of the patients were diagnosed with NSCLC for the first time and did not receive radiotherapy or chemotherapy before surgery. Our study was approved by the research ethics committee of the Tianjin Chest Hospital. Written informed consent was signed by all participants.
Cell Culture and Transfection
Five NSCLC cell lines (A549, H226, H292, ANP973 and H1299) (American Type Culture Collection, Manassas, ATCC, VA, USA) were cultured in RPMI-1640 medium containing 10% FBS. Normal human bronchial epithelial cell line BEAS-2B (ATCC) was cultured in BEGM medium (Lonza). All cell lines were incubated in a humidified incubator at 37°C with 5% CO2. Transfections were performed using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instruction. The siRNA sequences of SNHG6 were obtained from the study of Chang et al18 which had been proved to be effective. WIPF1 expression plasmid was purchased from ORIGENE (RC212019) and ETS1 siRNA was obtained from Santa Cruz Biotechnology (sc-37183).
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted using TRIzol (Invitrogen) and reverse-transcribed by the Prime Script TM RT Master Mix (TaKaRa Bio Technology). Quantitative PCR (qPCR) was performed using SYBR Premix Ex Taq TM (Takara). β-actin was used as an internal control for SNHG6, WIPF1, MMP2 and MMP9. The primers were as follows: SNHG6-F, 5ʹ-ATACTTCTGCTTCGTTACCT-3ʹ, SNHG6-R, 5ʹ-CTCATTTTCATCATTTGCT-3ʹ; ETS1-F,5ʹ-GATAGTTGTGATCGCCTCACC-3ʹ, ETS1-R, 5ʹ-GTCCTCTGAGTCGAAGCTGTC-3ʹ; WIPF1-F, 5ʹ-CGGAGGCGGTGGAAGTTTT-3ʹ, WIPF1-R, 5ʹ-CCGTGGATCTCAGCTTCGG-3ʹ; MMP2-F, 5ʹ-CCCACTGCGGTTTTCTCGAAT-3ʹ, MMP2-R, 5ʹ-CAAAGGGGTATCCATCGCCAT-3ʹ; MMP9-F, 5ʹ- AGACCTGGGCAGATTCCAAAC-3ʹ, MMP9-R, 5ʹ-CGGCAAGTCTTCCGAGTAGT-3ʹ; β-actin-F, 5ʹ-AGCGAGCATCCCCCAAAGTT-3ʹ, β-actin-R, 5ʹ-GGGCACGAAGGCTCATCATT-3ʹ. The TaqMan miRNA Reverse Transcription kit (Applied Biosystems) and TaqMan microRNA assay (Applied Biosystems) were used to analyze hsa-miR-944 and miR-181d-5p expression according to the manufacturer’s instructions. U6 snRNA was used as an internal control for miRNAs. The relative expression of genes was calculated using 2−ΔΔCT method.
Proliferation Assays
Cell proliferation was assessed using CCK-8 assay and cell cycle assay. Cells were transfected with siRNAs or miRNAs. At 24 h, 48 h and 72 h, CCK-8 reagents (Dojindo) were used to detect the relative proliferation activity. Cell cycle was analyzed with Cell Cycle and Apoptosis Analysis Kit (Beyotime, Shanghai, China) and Flow cytometry.
Migration Assays
Cell migration activity was assessed using wound-healing assay and transwell assay. Cells were cultured for 24 h and wounds were created with a 10μL pipette tip. Then, cells were cultured in DMEM without FBS at 37°C. Images were taken at 0 h and 24 h and the relative migration rates were analyzed. Transwell migration assays were conducted using 8-μm transwell inserts (Millipore). After 48 h of transfection, cells were digested and re-suspended in serum-free RPMI-1640 medium. Cells (2×104) were added into the upper chamber and RPMI-1640 medium with 10% FBS was added into the lower chamber. After culturing for 24 h, the cells on the upper side were removed and cells on the lower side were fixed with methanol and stained with 0.1% crystal violet.
Dual-Luciferase Reporter Assay
SNHG6 or ETS1 3ʹUTR sequences with the wild-type (WT) or mutated-type (MUT) of miR-944 or miR-181d-5p binding sites were cloned into pGLO vector (Promega). Cells were co-transfected with miRNA mimic and luciferase reporter plasmids and incubated for 48 h. Dual-Luciferase reporter assay system (Promega) was used to assess the luciferase activities.
The sequence 2kb upstream of WIPF1 CDS containing WT or MUT of ETS1 predicted binding site was inserted into a PGL3-Basic luciferase reporter (Promega). Cells were co-transfected with luciferase reporter and ETS1 expression plasmids and incubated for 48 h. Dual-Luciferase reporter assay system (Promega) was used to assess the luciferase activities.
RNA Immunoprecipitation
RNA immunoprecipitation (RIP) assays were performed with the RNA immunoprecipitation kit (Millipore, Billerica, MA, USA). Cells were collected and lysed with RIP lysis buffer and the extracts were incubated with Ago2 antibody-coated magnetic beads overnight at 4°C. The IgG antibody was taken as the negative control. The mixture was digested with protease K to isolate RNAs. qRT-PCR assays were applied to detect the abundance of SNHG6, miR-944 and miR-181d-5p.
Western Blot Assay
After 48 h of transfection, cells were collected to extract proteins using RIPA buffer. Proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred onto PVDF membranes. The membranes were then incubated with ETS1 primary antibodies (ab124282, Abcam), WIPF1 primary antibodies (sc-271114, Santa Cruz Biotechnology), MMP2 primary antibodies (ab97779, Abcam), MMP9 primary antibodies (ab38898, Abcam) and secondary antibodies. ECL immunoblotting kit was used to visualize the proteins.
Chromatin Immunoprecipitation (ChIP)
ChIP assay was performed with EZ ChIP kit (Millipore, Billerica, MA, USA) and ETS1 antibody (ab124282, Abcam). The primers used in the PCR analysis of this assay were: 5′-TGGTCAGAACCCCCTGATGA-3′ and 5′-TCCACAGCAAACATGTGGAGA-3′. Normal rabbit IgG (Beyotime, Shanghai, China) was used as a negative control in the immunoprecipitation.
Statistical Analyses
Data were analyzed using SPSS 20.0 software and p<0.05 was considered as statistically significant. The normality test was performed using Kolmogorov–Smirnov test. Student’s t-test or one-way ANOVA followed by Student-Newman-Keuls test were used for comparisons between groups. Data are presented as mean ± SD.
Results
SNHG6 Is Highly Expressed in NSCLC and Associated with Malignant Features of NSCLC
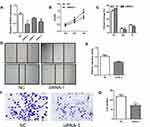
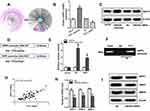
To investigate the role of SNHG6 in NSCLC, we first detected the expression level of SNHG6 in NSCLC tissues and cell lines using qRT-PCR. The results revealed that SNHG6 level was relatively higher in NSCLC tissues and cell lines compared with that in ANLs and BEAS-2B cell line (Figure 1A–C). To analyze the correlation between SNHG6 level and clinic-pathological features, NSCLC patients were divided into SNHG6 high-level group and SNHG6 low-level group according to the median. We found that SNHG6 expression was positively associated with TNM stage and tumor size (Table 1). Moreover, survival analysis suggested that high SNHG6 level displayed lower overall survival rate of NSCLC patients (Figure 1D).
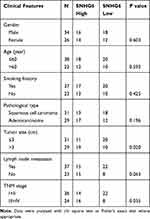 |
Table 1 Relationship Between SNHG6 Expression and Clinical Features of Patients with NSCLC |
Knockdown of SNHG6 Depresses the Proliferation and Migration of NSCLC Cells
To further explore the function of SNHG6 in NSCLC cells, we used siRNAs to reduce SNHG6 expression in A549 cells. The siRNA-1 exhibited the best efficiency in knockdown of SNHG6 in A549 cells, so we chose it in the following studies (Figure 2A). CCK8 assays and cell cycle assays showed that knockdown of SNHG6 depressed the proliferation (Figure 2B and C) and wound-healing assays and transwell assays reveal that knockdown of SNHG6 inhibited the migration (Figure 2D–G) of A549 cells. The above results indicated that SNHG6 might act as an oncogene to promote the progression of NSCLC.
miR-944 and miR-181d-5p are Targets of SNHG6
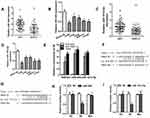
Through StarBase database, we found that SNHG6 contained putative complementary binding sites with some miRNAs. Among these miRNAs, miR-944 and miR-181d-5p caught our attention because they were reported to serve as tumor suppressor genes in NSCLC.19,20 First, we detected the expressions of miR-944 and miR-181d-5p in NSCLC tissues and cell lines. The results showed that miR-944 and miR-181d-5p levels were decreased in NSCLC tissues and cell lines (Figure 3A–D). Moreover, RIP assays showed that SNHG6, miR-944 and miR-181d-5p were significantly enriched in Ago2 conjugates (Figure 3E) which indicated that these genes may have a common acting basis. Then, we constructed luciferase reporter containing WT or MUT binding sites of SNHG6 sequence with miR-944 or miR-181d-5p (Figure 3F and G). Dual-luciferase reporter assay showed that miR-944 or miR-181d-5p only decreased the luciferase activity of WT SNHG6 luciferase reporter but not that of MUT SNHG6 luciferase reporter (Figure 3H and I). These results revealed that miR-944 and miR-181d-5p were targets of SNHG6.
SNHG6-miR-944/miR-181d-5p Axes are Involved in the Progression of NSCLC Cells
Given that SNHG6 could influence the activity of miR-944 and miR-181d-5p by binding to them, we then explored whether miR-944 and miR-181d-5p were involved in the progression inhibition induced by SNHG6 knockdown in NSCLC cells. CCK8 assays (Figure 4A), cell cycle assays (Figure 4B), wound-healing assays (Figure 4C and D) and transwell assays (Figure 4E and F) showed that miR-944 inhibitor or miR-181d-5p inhibitor reverses the inhibitory effects of SNHG6 siRNAs on A549 cells. These data suggested that SNHG6-miR-944/miR-181d-5p axis was involved in the progression of NSCLC cells.
SNHG6 Regulates ETS1 Expression via miR-944 and miR-181d-5p
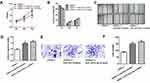
Through miRNA.org, we found ETS1 which was reported to be an oncogene in NSCLC21,22 might be a direct target of miR-944 (Figure 5A). Interestingly, ETS1 was also a potential target of miR-181d-5p (Figure 5B). Dual-luciferase reporter assay showed that miR-944 or miR-181d-5p only decreased the luciferase activity of WT ETS1 3ʹUTR luciferase reporter but not that of MT ETS1 3ʹUTR luciferase reporter (Figure 5C and D). Western blot analysis showed that miR-944 or miR-181d-5p could decrease the protein level of ETS1 (Figure 5E). These results revealed that ETS1 was a direct target of both miR-944 and miR-181d-5p. What’s more, knockdown of SNHG6 depressed the protein level of ETS1 and this effect could be reversed by miR-944 inhibitor or miR-181d-5p inhibitor (Figure 5F). These data altogether suggested that SNHG6 regulated ETS1 expression via miR-944 and miR-181d-5p.
ETS1 Enhances the Expression of WIPF1 via Binding to Its Promoter
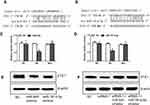
Using Coexpedia database, we did gene co-expression network analysis. The result showed that WIPF1 ranked in the first place of the potential genes co-expressed with ETS1 (Figure 6A). qRT-PCR assays and Western blot assays demonstrated that overexpression and knockdown of ETS1 significantly increased or decreased the expression of WIPF1 mRNA and protein expression (Figure 6B and C). Interestingly, ETS1 was found to have a binding region in the promoter of WIPF1 via JASPAR database (Figure 6D). Further experiments including dual-luciferase reporter assays and ChIP assays confirmed this interaction (Figure 6E and F). A strong correlation between ETS1 mRNA level and WIPF1 mRNA level in 60 NSCLC tissues was also discovered with qRT-PCR (Figure 6G). Therefore, we concluded that ETS1 enhanced the expression of WIPF1 via binding to its promoter. What’s more, both mRNA and protein level of ETS1 target genes including WIPF1, MMP223 and MMP924 could be restrained by SNHG6 silence (Figure 6H and I).
Discussion
In recent years, miRNAs and lncRNAs had been verified to play crucial roles in the development and progression of various cancers.25–27 In the current work, we investigated the role of lncRNA SNHG6 in NSCLC tumorigenesis and identified SNHG6 as an oncogenic factor in NSCLC.
First, we found SNHG6 was highly expressed in NSCLC tissues and cell lines. High SNHG6 level was correlated with malignant features in NSCLC such as advanced stage and large tumor size and poor prognosis of SCLC patients. The expression and clinical significance of SNHG6 had been reported in several cancers,15,28–30 which concluded that high SNHG6 level in cancers was associated with poor clinical outcome. Our study is the first study to show the expression and clinical significance of SNHG6 in NSCLC.
Additionally, we observed that SNHG6 knockdown could affect cell proliferation and migration of NSCLC cells in vitro. The oncogenic effect had already been described in some cancer but not in NSCLC.13–18 We also provided evidence that SNHG6 could bind to miR-944 and miR-181d-5p and affect their function. lncRNA–miRNA–mRNA competing endogenous RNA network has been widely recognized as a novel mode of regulation, in which lncRNAs may function as ceRNAs of mRNAs by competitively binding with their common miRNAs.31 The expression of ETS1 which we proved to be target of miR-944 and miR-181d-5p was thereby influenced by SNHG6. ETS1 belongs to the large family of the ETS domain family of transcription factors and primarily acts as a transcriptional activator. It is aberrantly expressed in many cancers and involved in cancer progression,32 and in NSCLC, ETS1 has been reported to be an oncogene to promote tumor progression.21,22 Furthermore, we demonstrated that ETS1 enhanced the expression of WIPF1 via binding to its promoter. WIPF1, WAS/WASL interacting protein family member 1, was verified to accelerate cancer progression in many cancers. For example, WIPF1 was reported to activate growth signals and increase YAP/TAZ and β-catenin stability, which thereby promote carcinogenic processes such as proliferation, stemness and invasiveness in glioma and breast cancer.33,34 WIPF1 can promote invasion, anchorage-independent growth and EMT progression via regulating RhoA in lung adenocarcinoma.35 Our data also revealed that SNHG6 could regulate the expression of ETS1 target genes including WIPF1, MMP2 and MMP9. MMP2 and MMP9 belong to matrix metalloproteinase (MMP) family which are well-known oncogenes in cancers and influencing cellular functions including growth, migration, invasion and gene expression.36–38 So we deem that SNHG6 may promote carcinogenesis via modulating ETS1 and its target genes.
Conclusion
In conclusion, our study illustrates that lncRNA SNHG6 is upregulated in NSCLC tissues and cell lines and its high expression is correlated with malignant features of NSCLC. SNHG6 facilitates carcinogenic progression by promoting the expressions of ETS1 and the downstream genes of ETS1 via miR-944 and miR-181d-5p, which may provide novel therapeutic strategies for NSCLC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sabour S. Prediction of post-operative morbidity and mortality in patients with lung cancer: methodological issues. Lung. 2018;196:499–500. doi:10.1007/s00408-018-0136-4
2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:2. doi:10.3322/caac.21262
3. Inage T, Nakajima T, Yoshino I, Yasufuku K. Early lung cancer detection. Clin Chest Med. 2018;39:45–55. doi:10.1016/j.ccm.2017.10.003
4. Criss SD, Mooradian MJ, Sheehan DF, et al. Cost-effectiveness and budgetary consequence analysis of Durvalumab consolidation therapy vs no consolidation therapy after chemoradiotherapy in stage III non-small cell lung cancer in the context of the US health care system. JAMA Oncol. 2018.
5. Qin J, Zeng N, Yang T, et al. Diagnostic value of autoantibodies in lung cancer: a systematic review and meta-analysis. Cell Physiol Biochem. 2018;51:2631–2646. doi:10.1159/000495935
6. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi:10.1016/j.cell.2018.01.011
7. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 1839;11:2014.
8. Xu T, Yan S, Jiang L, et al. Gene amplification-driven long noncoding RNA SNHG17 regulates cell proliferation and migration in human non-small-cell lung cancer. Mol Ther Nucleic Acids. 2019;17:405–413. doi:10.1016/j.omtn.2019.06.008
9. Wang W, Dong M, Zhang W, et al. Long noncoding LUCAT1 promotes cisplatin resistance of non-small cell lung cancer by promoting IGF-2. Eur Rev Med Pharmacol Sci. 2019;23:5229–5234. doi:10.26355/eurrev_201906_18188
10. Wang J, Cai H, Dai Z, et al. Down-regulation of lncRNA XIST inhibits cell proliferation via regulating miR-744/RING1 axis in non-small cell lung cancer. Clin Sci. 2019;133:1567–1579.
11. Shi X, Liu Z, Liu Z, et al. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:177–187. doi:10.1016/j.ebiom.2018.10.004
12. Yin D, Lu X, Su J, et al. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer. 2018;17:92. doi:10.1186/s12943-018-0836-7
13. Wang X, Lai Q, He J, et al. LncRNA SNHG6 promotes proliferation, invasion and migration in colorectal cancer cells by activating TGF-β/Smad signaling pathway via targeting UPF1 and inducing EMT via regulation of ZEB1. Int J Med Sci. 2019;16:51–59. doi:10.7150/ijms.27359
14. Wu Y, Deng Y, Guo Q, et al. Long non-coding RNA SNHG6 promotes cell proliferation and migration through sponging miR-4465 in ovarian clear cell carcinoma. J Cell Mol Med. 2019. doi:10.1111/jcmm.14359
15. Zhu X, Yang G, Xu J, et al. Silencing of SNHG6 induced cell autophagy by targeting miR-26a-5p/ULK1 signaling pathway in human osteosarcoma. Cancer Cell Int. 2019;19:82. doi:10.1186/s12935-019-0794-1
16. Jafari-Oliayi A, Asadi MH. SNHG6 is upregulated in primary breast cancers and promotes cell cycle progression in breast cancer-derived cell lines. Cell Oncol (Dordr). 2019;42:211–221. doi:10.1007/s13402-019-00422-6
17. Li Y, Li D, Zhao M, et al. Long noncoding RNA SNHG6 regulates p21 expression via activation of the JNK pathway and regulation of EZH2 in gastric cancer cells. Life Sci. 2018;208:295–304. doi:10.1016/j.lfs.2018.07.032
18. Chang L, Yuan Y, Li C, et al. Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR-101-3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Lett. 2016;383:183–194. doi:10.1016/j.canlet.2016.09.034
19. Liu M, Zhou K, Cao Y. MicroRNA-944 affects cell growth by targeting EPHA7 in non-small cell lung cancer. Int J Mol Sci. 2016;17:1493. doi:10.3390/ijms17101493
20. Gao L, Zheng Y, Wang P, et al. Tumor-suppressive effects of microRNA-181d-5p on non-small-cell lung cancer through the CDKN3-mediated Akt signaling pathway in vivo and in vitro. Am J Physiol Lung Cell Mol Physiol. 2019;316:L918–L933. doi:10.1152/ajplung.00334.2018
21. Zhou X, Zhou R, Zhou H, et al. ETS-1 induces endothelial-like differentiation and promotes metastasis in non-small cell lung cancer. Cell Physiol Biochem. 2018;45:1827–1839.
22. Cao B, Tan S, Tang H, et al. miR-512-5p suppresses proliferation, migration and invasion, and induces apoptosis in non-small cell lung cancer cells by targeting ETS1. Mol Med Rep. 2019;19:3604–3614. doi:10.3892/mmr.2019.10022
23. Higashikawa K, Yoneda S, Tobiume K, et al. DeltaNp63alpha-dependent expression of Id-3 distinctively suppresses the invasiveness of human squamous cell carcinoma. Int J Cancer. 2009;124:2837–2844. doi:10.1002/ijc.24280
24. Nazir SU, Kumar R, Singh A, et al. Breast cancer invasion and progression by MMP-9 through Ets-1 transcription factor. Gene. 2019;711:143952. doi:10.1016/j.gene.2019.143952
25. Lim LJ, Wong SYS, Huang F, et al. Roles and regulation of long non-coding RNAs in hepatocellular carcinoma. Cancer Res. 2019;79:5131–5139. doi:10.1158/0008-5472.CAN-19-0255
26. Rynkeviciene R, Simiene J, Strainiene E, et al. Non-coding RNAs in glioma. Cancers (Basel). 2018;11:17. doi:10.3390/cancers11010017
27. Panoutsopoulou K, Avgeris M, Scorilas A. miRNA and long non-coding RNA: molecular function and clinical value in breast and ovarian cancers. Expert Rev Mol Diagn. 2018;18:963–979. doi:10.1080/14737159.2018.1538794
28. Yan Y, Chen Z, Xiao Y, et al. Long non-coding RNA SNHG6 is upregulated in prostate cancer and predicts poor prognosis. Mol Biol Rep. 2019;46:2771–2778. doi:10.1007/s11033-019-04723-9
29. Yu C, Sun J, Leng X, et al. Long noncoding RNA SNHG6 functions as a competing endogenous RNA by sponging miR-181a-5p to regulate E2F5 expression in colorectal cancer. Cancer Manag Res. 2019;11:611–624. doi:10.2147/CMAR
30. Lv P, Qiu X, Gu Y, et al. Long non-coding RNA SNHG6 enhances cell proliferation, migration and invasion by regulating miR-26a-5p/MAPK6 in breast cancer. Biomed Pharmacother. 2019;110:294–301. doi:10.1016/j.biopha.2018.11.016
31. Wang Y, Huang T, Sun X, et al. Identification of a potential prognostic lncRNA-miRNA-mRNA signature in endometrial cancer based on the competing endogenous RNA network. J Cell Biochem. 2019;120:18845-18853.
32. Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol. 2015;35:20–38. doi:10.1016/j.semcancer.2015.09.010
33. Rivas S, Antón IM, Wandosell F. WIP-YAP/TAZ as A new pro-oncogenic pathway in glioma. Cancers (Basel). 2018;10:191.
34. Escoll M, Gargini R, Cuadrado A, et al. Mutant p53 oncogenic functions in cancer stem cells are regulated by WIP through YAP/TAZ. Oncogene. 2017;36:3515–3527. doi:10.1038/onc.2016.518
35. Salvi A, Thanabalu T. WIP promotes in-vitro invasion ability, anchorage independent growth and EMT progression of A549 lung adenocarcinoma cells by regulating RhoA levels. Biochem Biophys Res Commun. 2017;482:1353–1359. doi:10.1016/j.bbrc.2016.12.040
36. Tauro M, Lynch CC. Cutting to the chase: how matrix metalloproteinase-2 activity controls breast-cancer-to-bone metastasis. Cancers (Basel). 2018;10(6):185. doi:10.3390/cancers10060185
37. Thakur V, Bedogni B. The membrane tethered matrix metalloproteinase MT1-MMP at the forefront of melanoma cell invasion and metastasis. Pharmacol Res. 2016;111:17–22. doi:10.1016/j.phrs.2016.05.019
38. Roy R, Yang J, Moses MA. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J Clin Oncol. 2009;27:5287–5297. doi:10.1200/JCO.2009.23.5556
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.