Back to Journals » OncoTargets and Therapy » Volume 13
Long Noncoding RNA RMRP Suppresses the Tumorigenesis of Hepatocellular Carcinoma Through Targeting microRNA-766
Authors Shao C, Liu G, Zhang X, Li A, Guo X
Received 25 December 2019
Accepted for publication 17 March 2020
Published 8 April 2020 Volume 2020:13 Pages 3013—3024
DOI https://doi.org/10.2147/OTT.S243736
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Cunhua Shao,1 Gongpan Liu,1 Xiaobin Zhang,1 Anyun Li,2 Xingjun Guo1
1Department of Hepatobiliary Surgery, Dongying People’s Hospital, Dongying City 257091, People’s Republic of China; 2Department of General Surgery, Dongying Hong Gang Hospital, Dongying City 257000, People’s Republic of China
Correspondence: Xingjun Guo
Department of Hepatobiliary Surgery, Dongying People’s Hospital, No. 317, Nanyi Road, Dongying City 257091, People’s Republic of China
Tel +86 0546 8901126
Email [email protected]
Purpose: This study aimed to explore the regulatory effect of long noncoding RNA (lncRNA) ribonuclease mitochondrial RNA processing gene (RMRP) on hepatocellular carcinoma (HCC).
Methods: The expression of RMRP in HCC tissues and cell lines was assessed by qRT-PCR. Kaplan–Meier method was utilized to analyze the correlation between RMRP expression and the survival of HCC patients. MHCC97H and HuH7 cells were transfected with pcDNA3.1-RMRP or pcDNA3.1, respectively. MTT and flow cytometry assays were conducted to examine the proliferation and apoptosis of HCC cells, respectively. The migration and invasion of HCC cells were assessed using wound healing and transwell assays, respectively. StarBase3.0 and dual-luciferase reporter gene assay were used to identify the target relationship between miR-766 and RMRP. A xenografted tumor model was established in rats to evaluate the effect of RMRP in vivo.
Results: RMRP was down-regulated in HCC tissues and cells. Low expression of RMRP was correlated with poor survival of HCC patients. The A495 value and colony number were significantly decreased in pcDNA3.1-RMRP-transfected MHCC97H and HuH7 cells. The apoptosis rate was significantly increased in pcDNA3.1-RMRP-transfected MHCC97H and HuH7 cells. The migration rate and the number of invasive cells were significantly decreased in pcDNA3.1-RMRP-transfected MHCC97H and HuH7 cells. MiR-766 was a target of RMRP and eliminated the anti-tumor effect of RMRP on MHCC97H cells. The up-regulation of RMRP suppressed the growth of xenograft tumors in rats.
Conclusion: Overexpression of RMRP suppressed the tumorigenesis of HCC by targeting miR-766.
Keywords: LncRNA RMRP, miR-766, hepatocellular carcinoma, proliferation, migration, invasion
Introduction
Hepatocellular carcinoma (HCC) is the most prevalent subtype of liver cancer, accounting for 70–85% of liver cancer in the world.1,2 In recent years, surgical resection is still the predominant therapeutic strategy for HCC, while it does not has a desirable outcome with an overall survival rate of approximately 50% within 5 years.3,4 The most effective therapeutic method for early stage HCC is liver transplantation or curative surgery, while therapeutic options for advanced HCC are very limited.5,6 Therefore, exploring novel therapeutic targets is urgently needed to ameliorate the prognosis of HCC patients.7–9
Long noncoding RNAs (lncRNAs) are a major class of noncoding RNA transcripts with length over 200 nucleotides.10 LncRNAs are involved in diverse biological processes, such as genomic imprinting, epigenetic regulation, and the regulation of cell cycle, apoptosis, and differentiation, via interacting with DNA, RNA molecules or proteins.11 Recently, many researches have suggested that lncRNAs play a critical role in the occurrence and metastasis of cancers.12,13 For example, lncRNA HULC accelerates the tumor invasion and metastasis of HCC.14,15 LncRNA UCA1 is up-regulated in HCC tissues, and UCA1 depletion represses the metastasis of HCC.16 LncRNA RNA component of mitochondrial RNA processing endoribonuclease (RMRP) is widely expressed in murine and human tissues.17 Meng et al18 have demonstrated that ectopic expression of RMRP promotes the proliferation and invasion of lung adenocarcinoma cells. Feng et al19 have indicated that RMRP serves as an oncogene and a therapeutic target for glioma. Wang et al20 have revealed that RMRP accelerates the proliferation and invasion of non-small cell lung cancer (NSCLC) cells through targeting miR‐1‐3p. However, the knowledge on the regulatory effect and mechanism of RMRP on HCC remains limited.
LncRNAs are involved in the development and progression of various types of cancers through binding to specific microRNAs (miRNAs).21 For example, lncRNA MIR4435-2HG accelerates the proliferation of HCC cells through up-regulating miR-487a.22 LncRNA DGCR5 suppresses the progression of HCC through targeting miR-346.23 MiRNAs play key roles in the pathogenesis of HCC, and a large number of miRNAs are dysregulated in HCC, such as miR-1, 21, −25, −26a, −375, −206, −223, −224, −92a, −222, and -let-7f.24–28 MiR-766 is a novel defined metastasis-related miRNA in human, which is identified through the comparison between a primary xenograft model and a metastasis model.29 Increasing researches have demonstrated that miR-766 serves as either a tumor-promoter or a tumor-suppressor in different cancers, such as renal cell carcinoma,30 colorectal cancer,31 breast cancer,32 and lung cancer.33 Chen et al have proved that DNA methylation-modulated suppression of miR-766-3p accelerates the proliferation of renal cell carcinoma cells.30 Bai et al33 have indicated that lncRNA CASC15 accelerates the proliferation and invasion of lung cancer cells via modulating miR-766-5p/KLK12 axis. However, the regulatory relationship between RMRP and miR-766 in HCC remains unclear.
In the present study, we first detected the expression of RMRP in HCC tissues, and the effects of RMRP overexpression on the proliferation, migration, invasion and apoptosis of HCC cells. The regulatory relationship between miR-766 and RMRP was then predicted and confirmed. A xenografted tumor model was established in rats to analyze the anti-tumor effect of RMRP on HCC in vivo. Our research revealed the regulatory effect of RMRP on the development of HCC and provided a potential therapeutic target for HCC.
Materials and Methods
Clinical Specimens
Totally 40 tumor tissues and adjacent normal tissues were obtained from HCC patients (28 males, 12 females) receiving surgical resection from April 2013 to May 2014. HCC tissues were histopathologically confirmed. These patients did not receive preoperative adjuvant chemotherapy, radiotherapy, targeted therapy or immunotherapy before surgical resection. This study was permitted by the Ethics Committee of Dongying People’s Hospital and written informed consents were obtained from all patients.
Cell Culture
HCC cell lines (Hep3B, HepG2, MHCC97H and HuH7) and human hepatic cell line HL-7702 (L02) were obtained from Cell Bank of Type Culture Collection (CBTCC, Shanghai, China). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Waltham, MA, USA) and maintained in an incubator (MCO-15AC, Sanyo, JAPAN) at 37°C with 5% CO2. When growing to 80% confluence, cells were digested with 0.25% trypsin and then passaged every other day.
Cell Transfection
Cells were seeded into 6-well cell plates (6×105 cells/well) and cultured at 37°C overnight. Transfection was conducted using Lipofectamine 2000 (Invitrogen, Waltham, MA, USA) following manufacturer’s instructions. MHCC97H and HuH7 cells were transfected with pcDNA3.1-RMRP or pcDNA3.1 negative control, and grouped as the pcDNA3.1-RMRP or pcDNA3.1-NC group, respectively. MHCC97H cells were further co-transfected with miR-766 mimics or mimics-NC (GenePharma Co., Ltd, Shanghai, China) and pcDNA3.1-RMRP or pcDNA3.1-NC, and grouped as the mimics-NC, miR-766 mimics, NC + mimics-NC, RMRP + mimics-NC, NC + miR-766 mimics, and RMRP + miR-766 mimics group. Cells without transfection were considered as the Mock group.
qRT-PCR
Trizol reagent (Invitrogen) was used to isolate the total RNA from cells in each group. RNA reverse transcription was conducted using reverse transcription kit (Takara, Otsu, Japan). qRT-PCR was conducted on ABI 7500HT Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) under the following reaction conditions: 95°C for 10 min, 40 cycles at 95°C for 15 s, 60°C for 30 s and 72°C for 20 s. The expression level of mRNA was calculated according to the 2−ΔΔCt method.34 The primer sequences are shown in Table 1. U6 or β-actin was used as the internal reference of miR-766 or RMRP, respectively.
 |
Table 1 Primer Sequences |
MTT Analysis
Cell viability was assessed by MTT assay as previously reported.35 Cells were seeded in 96-well plates (6 ×103 cells/well) and cultured for 0, 24, 48, and 72 h, respectively. Subsequently, 20 μL MTT (5 mg/mL, Sigma-Aldrich, St. Louis, MO, USA) was pipetted into each well. After 4 h of incubation, 150 μL DMSO was added to terminate the reaction. The absorbance at 495 nm (A495) was measured by a microplate reader (Applied Biosystems).
Colony Formation Assay
Cell proliferation was assessed by colony formation assay as previously reported.36 After transfected for 48 h, cells were seeded in 6-well plates (600 cells/well, 2.5 mL medium/well) and cultured for 14 d. Cells were then stained with crystal violet for 15 min. The stained colonies (more than 50 cells) were photographed under a microscope (OlympusCkx53, Japan) and were counted using ImageJ (1.48V) software.
AnnexinV-PI Double Staining
Cell proliferation was assessed by AnnexinV-PI double staining as previously reported.37 A total of 1×105 cells were suspended in 500 µL binding buffer. Cells were then stained with 5 µL Annexin V-EGFP and 5mL Propidium Iodide (Invitrogen). Cell apoptosis rate was examined by a MUSETM flow cytometer (Merck Millipore, USA).
Wound Healing Assay
Cell migration was assessed by wound healing assay as previously reported.38 Cells were seeded into 6-well plates (5×105 cells/well). When cells were grown to 90% confluence, an artificial scratch was created using a 10 μL pipette tip. Cells were then cultured for 24 h and observed under an inverted microscope (Olympus). Wound healing rate was calculated by the fraction of cell coverage across the line.
Transwell Assay
Cell invasion was assessed using transwell membranes (BD, Franklin Lakes, NJ, USA) coated with Matrigel as previously reported.38 Totally 100 μL cells (1×105/100 μL) were seeded into the upper chamber, and 500 μL medium containing 10% FBS was added to the lower chamber as a chemoattractant. After 48 h of incubation, the invasive cells were stained with crystal violet for 10 min. Positive stained cells were counted under a microscope (Olympus) at five random fields.
Dual-Luciferase Reporter Gene Assay
The miRNA targets of RMRP were predicated by StarBase3.0. MiR-766 was chosen because of high rank and its tumor-promoting role in HCC.39 A binding site of RMRP on miR-766 was predicated. According to the predication, the fragments containing the binding site and the mutant site were cloned into pmirGLO vector (Promega, Madison, WI, USA), and separately named RMRP-Wt and RMRP-Mut. MHCC97H cells were then co-transfected with RMRP-Mut or RMRP-Wt and miR-766 mimics or miR-766-NC (GenePharma Co., Ltd) using Lipofectamine 3000 (L3000015, Thermo Fisher Scientific, Waltham, MA, USA). After transfected for 48 h, dual-luciferase reporter gene assay system (Promega) was used to detect the luciferase activity.40
Xenografted Tumor Model
Male nude rats (BALB/c) at 4 weeks old were obtained from Shanghai experimental animal center, Chinese academy of sciences (Shanghai, China). Rats were randomly divided into two groups (5 rats in each group). MHCC97H cells at the logarithmic growth phase were collected and subcutaneous injected into the left axilla of rats (1×107cells/rat). The longest diameter (L) and the shortest diameter (W) of the tumor were measured with vernier caliper every 7 days after injection. Tumor volume was calculated using the following formula: V=L×W2/2. At the end of the 4th week, the tumor was completely dissected and weighted.41 All animal experimental procedures were permitted by the Ethics Committee of Dongying People’s Hospital, and were performed in accordance with the Guide for the Care and Use of Laboratory Animals (eighth edition, 2011, National Institutes of Health, USA).
Statistical Analysis
All assays were conducted for at least three times. Data were analyzed by SPSS 22.0 statistical software and GraphPad Prism 7.0. Data were presented as mean ± standard deviation (SD). Student’s t-test was used to compare the significant difference of two groups. One-way ANOVA followed by Tukey’s post hoc test was applied to compare the significant difference of more than two groups. Difference was regarded statistically significant at P < 0.05.
Results
The Expression of RMRP Was Decreased in HCC Tissues
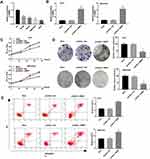
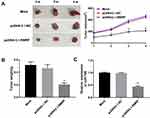
As presented in Figure 1A, the expression of RMRP in tumor tissues was significantly lower than that in normal tissues (P < 0.0001). Patients with HCC were classified into high and low expression groups according to the median expression of RMRP. The correlation between the RMRP expression and the clinicopathological features is shown in Table 2. The down-regulation of RMRP was significantly associated with the TNM stage, tumor size, and metastasis (P < 0.05). In addition, Kaplan-Meier survival assay showed that the 5-year survival rate of patients in the high RMRP expression group was 29% and the median survival time was 40 months. The 5-year survival rate of patients in the low RMRP expression group was 16% and the median survival time was 17 months (P = 0.0047, Figure 1B).
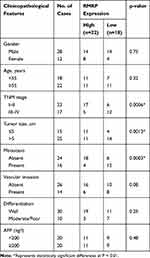 |
Table 2 Relation Between RMRP Expression and Clinicopathological Features in HCC |
Up-Regulation of RMRP Inhibited the Proliferation and Induced the Apoptosis of HCC Cells
Compared with normal human hepatic cell line L02, the expression of RMRP was significantly decreased in Hep3B, HepG2, MHCC97H and HuH7 cells (all P < 0.01, Figure 2A). MHCC97H and HuH7 cells with relatively low expression of RMRP were used in subsequent assays. The expression of RMRP expression significantly up-regulated in the pcDNA3.1-RMRP group compared with that in the Mock group (P < 0.01, Figure 2B). MTT and colony formation assay showed that the A495 value and the colony number were significantly lower in the pcDNA3.1-RMRP group than that in the Mock group (P < 0.05, Figure 2C and D). In addition, the apoptosis rate was significantly increased in the pcDNA3.1-RMRP group compared to the Mock group (Figure 2E, P < 0.01). There was no significant difference between the pcDNA3.1-NC group and the Mock group.
Up-Regulation of RMRP Inhibited the Migration and Invasion of HCC Cells
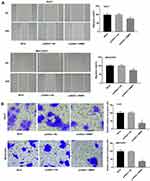
Wound healing and transwell assay showed that the migration rate and the relative number of invasive cells were markedly decreased in the pcDNA3.1-RMRP group compared with the Mock group (P < 0.01, Figure 3A and B). There was no significant change in the pcDNA3.1-NC group compared with the Mock group.
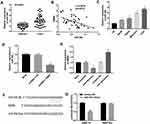
MiR-766 Was the Target of RMRP
The expression of miR-766 in tumor tissues was significantly higher than that in normal tissues (P < 0.0001, Figure 4A). Spearman correlation assay showed that the expression of miR-766 was negatively correlated with the expression of RMRP in tumor tissues (r =﹣0.4939, P =0.0012, Figure 4B). In addition, the expression of miR-766 was significantly higher in HCC cell lines (Hep3B, HepG2, MHCC97H and HuH7) than that in normal hepatocyte cell line L02 (P < 0.01, Figure 4C). As presented in Figure 4D, the expression of miR-766 was significantly decreased in the pcDNA3.1-RMRP group compared with that in the Mock group (P < 0.05, Figure 4D). To investigate the regulatory effect of miR-766 on RMRP, miR-766 mimics or miR-766 inhibitor was transfected into MHCC97H cells. The expression of RMRP was significantly decreased in the miR-766 mimics group compared with the mimics-NC group, and was significantly increased in the miR-766 inhibitor group compared with the inhibitor-NC group (P < 0.05, Figure 4E). Furthermore, StarBase3.0 predicted a binding site of RMRP on miR-766 (Figure 4F). Dual-luciferase reporter gene assay showed that the co-transfection of miR-766 mimics and RMRP Wt significantly decreased the luciferase activity of MHCC97H cells, and co-transfection of miR-766 mimics and RMRP Mut did not influence the luciferase activity (P < 0.01, Figure 4G). The above results revealed that miR-766 was a target of RMRP.
MiR-766 Eliminated the Effect of RMRP on HCC Cells
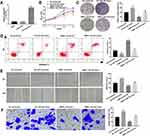
To investigate the regulatory relationship between miR-766 and RMRP on HCC cells, miR-766 mimics were transfected into MHCC97H cells. The expression of miR-766 in the miR-766 mimics group was significantly increased compared with the Mock group (P < 0.01, Figure 5A). The miR-766 mimics and pcDNA3.1-RMRP were then co-transfected into MHCC97H cells. As shown in Figure 5C, E and F, the cell proliferation, migration and invasion were significantly enhanced in the NC + miR-766 mimics group, and were significantly inhibited in the RMRP + mimics-NC group compared with the NC + mimics-NC group (P < 0.01). The results of cell apoptosis were opposite to those of cell proliferation (P < 0.01, Figure 5D). Notably, miR-766 mimics eliminated the anti-tumor effects of RMRP on HCC cells (P < 0.05, Figure 5B–F).
Up-Regulation of RMRP Repressed the Xenograft Tumor Growth in Rats
To further evaluate the anti-tumor effect of RMRP on HCC in vivo, xenograft tumor model was established in rats. The tumor volume and weight were significantly decreased in the pcDNA3.1-RMRP group compared with those in the pcDNA3.1-NC group (P < 0.05, Figure 6A and B). qRT-PCR showed that the expression of miR-766 in the pcDNA3.1-RMRP group was significantly lower than that in the pcDNA3.1-NC group (P < 0.01, Figure 6C). The above results indicated that the up-regulation of RMRP inhibited the xenograft tumor growth in rats by down-regulating miR-766.
Discussion
HCC is a type of malignant tumor with poor prognosis and high mortality worldwide.42 Over the past ten years, HCC has become one of the most frequently occurring cancers. There about one-third of cancer-related deaths are caused by HCC around the world.43 The high recurrence of HCC leads to a poor prognosis, which promotes the exploration of novel therapeutic targets for HCC.
The deregulation of lncRNAs plays a key role in the development of HCC.44 Many lncRNAs have been identified to be down-regulated in HCC, such as 00364,45 DGCR5,46 MEG3,47 and RP1130-1.48 In consistent with these lncRNAs, RMRP was significantly down-regulated in HCC tissues and cells in this study. Previous studies have confirmed that some lncRNAs are potential prognostic markers for HCC. Huang et al have shown that the low expression of DGCR5 is correlated with a poor survival of patients with HCC.46 Xiao et al have proved that the low expression of RP1130-1 is associated with the clinical stage, tumor number, microvascular invasion, and short recurrence-free survival in HCC.48 In this study, the down-regulation of RMRP was significantly associated with the TNM stage, tumor size, metastasis, and poor survival in patients with HCC. These findings indicate that RMRP is a potential prognostic marker for HCC.
The roles of lncRNAs on cancers are mainly reflected in the regulation of cellular biological processes, such as cell proliferation, apoptosis, migration and invasion. Some lncRNAs have been considered to be potential therapeutic targets for HCC by regulating these processes. Tang et al have shown that the overexpression of 00364 represses the proliferation and induces the apoptosis of HCC cells.45 Mo et al have proved that the overexpression of FAM99B significantly inhibits the proliferation, migration, and invasion of HCC cells.49 Zhuang et al have demonstrated that DRHC suppresses the proliferation, migration, and invasion of HCC cells.50 In this study, we found that the up-regulation of RMRP inhibited the proliferation, migration and invasion, and induced the apoptosis of HCC cells. These results illustrate that RMRP is a tumor suppressor in HCC cells. RMRP exhibits a similar anti-tumor effect on HCC with the lncRNAs mentioned above. To further identify the anti-tumor role of RMRP in vivo, a xenografted tumor model was established in rats. We found that the overexpression of RMRP repressed the xenograft tumor growth in rats. These results further confirmed that RMRP can inhibit the tumorigenesis of HCC in vivo.
MiRNAs are usually acted as the regulatory targets of lncRNAs that involved in the development of human cancers. In this study, a binding site of RMRP on miR-766 was predicted by StarBase3.0. MiR-766 was further identified as a target of RMRP by dual-luciferase reporter gene assay. MiR-766 is known as a human metastasis-related miRNA that is up-regulated in diverse types of cancers, including colorectal cancer,51 cutaneous squamous cell carcinoma,52 lung adenocarcinoma53 and acute promyelocytic leukemia.54 In consistent with previous researches, the expression of miR-766 was significantly up-regulated in HCC tissues and cells in this study. Li et al have found that the ectopic expression of miR-766 promotes cell growth and anchorage-independent growth of colorectal cancer cells.51 Because RMRP could negatively regulated miR-766 in HCC cells, we speculate that RMRP may inhibit the tumorigenesis of HCC via down-regulating miR-766. This speculation was further confirmed by our following feedback assays. The results showed that miR-766 overexpression eliminated the anti-tumor effect of RMRP on HCC cells. Our findings demonstrate that RMRP can repress the proliferation, migration and invasion of HCC cells via targeting miR-766.
Conclusions
In conclusion, RMRP was down-regulated in HCC tissues and correlated with the poor prognosis of HCC patients. Overexpression of RMRP inhibited the proliferation, migration and invasion, and promoted the apoptosis of HCC cells via targeting miR-766. Overexpression of RMRP also repressed the xenograft tumor growth in rats. RMRP may serve as a potential prognostic biomarker and a therapeutic target for HCC.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Ethics and Consent Statement
This study was permitted by the Ethics Committee of Dongying People’s Hospital and written informed consents were obtained from all patients. All animal experimental procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (eighth edition, 2011, National Institutes of Health, USA).
Disclosure
Cunhua Shao and Gongpan Liu are co-first authors. The authors report no conflicts of interest in this work.
References
1. Schmieder R, Puehler F, Neuhaus R, et al. Allosteric MEK1/2 inhibitor refametinib (BAY 86-9766) in combination with sorafenib exhibits antitumor activity in preclinical murine and rat models of hepatocellular carcinoma. Neoplasia. 2013;15(10):1147–1157. doi:10.1593/neo.13812
2. Su JC, Tseng PH, Wu SH, et al. SC-2001 overcomes STAT3-mediated sorafenib resistance through RFX-1/SHP-1 activation in hepatocellular carcinoma. Neoplasia. 2014;16(7):595–605. doi:10.1016/j.neo.2014.06.005
3. El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. doi:10.1053/j.gastro.2007.04.061
4. Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):
5. Llovet JM. Liver cancer: time to evolve trial design after everolimus failure. Nat Rev Clin Oncol. 2014;11(9):506. doi:10.1038/nrclinonc.2014.136
6. Singal A, Volk ML, Waljee A, et al. Meta-analysis: surveillance with ultrasound for early-stage hepatocellular carcinoma in patients with cirrhosis. Aliment Pharmacol Ther. 2010;30(1):37–47. doi:10.1111/j.1365-2036.2009.04014.x
7. Colecchia A, Schiumerini R, Cucchetti A, et al. Prognostic factors for hepatocellular carcinoma recurrence. World J Gastroenterol. 2014;20(20):5935–5950. doi:10.3748/wjg.v20.i20.5935
8. Lee SC, Tan HT, Chung MCM. Prognostic biomarkers for prediction of recurrence of hepatocellular carcinoma: current status and future prospects. World J Gastroenterol. 2014;20(12):3112–3124. doi:10.3748/wjg.v20.i12.3112
9. Naeli P, Pourhanifeh MH, Karimzadeh MR, et al. Circular RNAs and gastrointestinal cancers: epigenetic regulators with a prognostic and therapeutic role. Crit Rev Oncol Hematol. 2020;145:102854. doi:10.1016/j.critrevonc.2019.102854
10. Wilusz JE, Hongjae S, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi:10.1101/gad.1800909
11. Vafadar A, Shabaninejad Z, Movahedpour A, et al. Long non-coding RNAs as epigenetic regulators in cancer. Curr Pharm Des. 2019;25(33):3563–3577. doi:10.2174/1381612825666190830161528
12. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
13. Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: a potential novel class of cancer biomarkers. Front Genet. 2015;6:145. doi:10.3389/fgene.2015.00145
14. Hui X, Hongwei M, Danqiu Z. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013(23):1–5.
15. Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–342. doi:10.1053/j.gastro.2006.08.026
16. Wang F, Ying HQ, He BS, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6(10):7899.
17. Rosenbluh J, Nijhawan D, Chen Z, Wong KK, Masutomi K, Hahn WC. RMRP is a non-coding RNA essential for early murine development. PLoS One. 2011;6(10):e26270. doi:10.1371/journal.pone.0026270
18. Meng Q, Ren M, Li Y, Song X. LncRNA-RMRP acts as an oncogene in lung cancer. PLoS One. 2016;11(12):e0164845. doi:10.1371/journal.pone.0164845
19. Feng W, Li L, Xu X, Jiao Y, Wei D. Up-regulation of the long non-coding RNA RMRP contributes to glioma progression and promotes glioma cell proliferation and invasion. Arch Med Sci. 2017;13(6):1315–1321. doi:10.5114/aoms.2017.66747
20. Wang Y, Luo X, Liu Y, Han G, Sun D. Long noncoding RNA RMRP promotes proliferation and invasion via targeting miR. J Cell Biochem. 2019;120(9):15572–15584. doi:10.1002/jcb.28824
21. Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;35(3):507. doi:10.1093/carcin/bgt405
22. Kong Q, Liang C, Jin Y, et al. The lncRNA MIR4435-2HG is upregulated in hepatocellular carcinoma and promotes cancer cell proliferation by upregulating miRNA-487a. Cell Mol Biol Lett. 2019;24(1). doi:10.1186/s11658-019-0148-y.
23. Wang YG, Liu J, Shi M, Chen FX. LncRNA DGCR5 represses the development of hepatocellular carcinoma by targeting the miR-346/KLF14 axis. J Cell Physiol. 2018;234(1):572–580. doi:10.1002/jcp.26779
24. Mirzaei HR, Sahebkar A, Mohammadi M, et al. Circulating microRNAs in hepatocellular carcinoma: potential diagnostic and prognostic biomarkers. Curr Pharm Des. 2016;22(34):5257–5269. doi:10.2174/1381612822666160303110838
25. Gong J, He XX, Tian A. Emerging role of microRNA in hepatocellular carcinoma (Review). Oncol Lett. 2015;9(3):1027–1033. doi:10.3892/ol.2014.2816
26. Zhou L, Yang X, Song W-J, et al. MicroRNA-21 regulates the migration and invasion of a stem-like population in hepatocellular carcinoma. Int J Oncol. 2013;43(2):661–669. doi:10.3892/ijo.2013.1965
27. Yu W, Lee ATC, Ma JZI, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target *. J Biol Chem. 2008;283(19):13205–13215. doi:10.1074/jbc.M707629200
28. Xin Y, Lei L, Xiao-Fei Z, et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58(1):158–170. doi:10.1002/hep.26305
29. Hyeong-Gon M, Keunhee O, Jiwoo L, et al. Prognostic and functional importance of the engraftment-associated genes in the patient-derived xenograft models of triple-negative breast cancers. Breast Cancer Res Treat. 2015;154(1):1–10.
30. Chen C, Xue S, Zhang J, et al. DNA-methylation-mediated repression of miR-766-3p promotes cell proliferation via targeting SF2 expression in renal cell carcinoma. Int J Cancer. 2017;141(9):1867. doi:10.1002/ijc.30853
31. Afgar A, Fard-Esfahani P, Mehrtash A, et al. MiR-339 and especially miR-766 reactivate the expression of tumor suppressor genes in colorectal cancer cell lines through DNA methyltransferase 3B gene inhibition. Cancer Biol Ther. 2016;17(11):1126–1138. doi:10.1080/15384047.2016.1235657
32. Oh K, Lee DS. In vivo validation of metastasis-regulating microRNA-766 in human triple-negative breast cancer cells. Lab Anim Res. 2017;33(3):256–263. doi:10.5625/lar.2017.33.3.256
33. Bai Y, Zhang G, Cheng R, Yang R, Chu H. CASC15 contributes to proliferation and invasion through regulating miR-766-5p/ KLK12 axis in lung cancer.Cell Cycle. 2019;18(18):2323–2331. doi:10.1080/15384101.2019.1646562.
34. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
35. Li J, Jia L, Ma ZH, Ma QH, Zhao YF. Axl glycosylation mediates tumor cell proliferation, invasion and lymphatic metastasis in murine hepatocellular carcinoma. World J Gastroenterol. 2012;18(38):5369–5376. doi:10.3748/wjg.v18.i38.5369
36. Ge H, Chaojie L, Zhixia L, et al. DcR3 induces proliferation, migration, invasion, and EMT in gastric cancer cells via the PI3K/AKT/GSK-3β/β-catenin signaling pathway. OncoTargets Ther. 2018;11:4177–4187. doi:10.2147/OTT
37. Zhang Z, Wang J, Li J, Wang X, Song W. MicroRNA-150 promotes cell proliferation, migration, and invasion of cervical cancer through targeting PDCD4. Biomed Pharmacother. 2017;97:511–517. doi:10.1016/j.biopha.2017.09.143
38. Jin Y, Feng SJ, Qiu S, Shao N, Zheng JH. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2017;21(14):3176–3184.
39. Yang C, Ma X, Guan G, et al. MicroRNA-766 promotes cancer progression by targeting NR3C2 in hepatocellular carcinoma. FASEB J. 2019;33(1):1456–1467. doi:10.1096/fsb2.v33.1
40. Fu X, Wen H, Jing L, et al. MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci. 2017;108(4):620–631. doi:10.1111/cas.13177
41. Xiao ZQ, Teng-Kun Y, Ya-Xing L, Jian-He Z, Jian-Jun G. miR-130b regulates the proliferation, invasion and apoptosis of glioma cells via targeting of CYLD. Oncol Rep. 2017;38(1):167–174. doi:10.3892/or.2017.5651.
42. Melchiorre C, Mccubrey JA, Antonella C, Nadia L, Antonina A, Giuseppe M. Targeted therapy for hepatocellular carcinoma: novel agents on the horizon. Oncotarget. 2012;3(3):236–260. doi:10.18632/oncotarget.466
43. Han LL, Lv Y, Guo H, Ruan ZP, Nan KJ. Implications of biomarkers in human hepatocellular carcinoma pathogenesis and therapy. World J Gastroenterol. 2014;20(30):10249. doi:10.3748/wjg.v20.i30.10249
44. Zhu J, Liu S, Ye F, et al. The long noncoding RNA expression profile of hepatocellular carcinoma identified by microarray analysis. PLoS One. 2014;9(7):e101707. doi:10.1371/journal.pone.0101707
45. Tang WG, Hu B, Sun HX, et al. Long non-coding RNA00364 represses hepatocellular carcinoma cell proliferation via modulating p-STAT3-IFIT2 signaling axis. Oncotarget. 2017;8(60):102006–102019. doi:10.18632/oncotarget.22039
46. Huang R, Wang X, Zhang W, et al. Down-regulation of LncRNA DGCR5 correlates with poor prognosis in hepatocellular carcinoma. Cell Physiol Biochem. 2016;40(3–4):707–715. doi:10.1159/000452582
47. He JH, Han ZP, Liu JM, Zhou JB, Cao MR. Overexpression of long non-coding RNA MEG3 inhibits proliferation of hepatocellular carcinoma Huh7 cells via negative modulation of miRNA-664. J Cell Biochem. 2017;118(11):3713–3721. doi:10.1002/jcb.v118.11
48. Xiao C, Wang C, Cheng S, et al. The significance of low levels of LINC RP1130-1 expression in human hepatocellular carcinoma. Biosci Trends. 2016;10(5):378–385. doi:10.5582/bst.2016.01123
49. Mo M, Liu S, Ma X, et al. A liver-specific lncRNA, FAM99B, suppresses hepatocellular carcinoma progression through inhibition of cell proliferation, migration, and invasion. J Cancer Res Clin Oncol. 2019;145(8):2027–2038. doi:10.1007/s00432-019-02954-8
50. Zhuang R, Zhang X, Lu D, et al. lncRNA DRHC inhibits proliferation and invasion in hepatocellular carcinoma via c‐Myb‐regulated MEK/ERK signaling. Mol Carcinog. 2019;58(3):366–375. doi:10.1002/mc.v58.3
51. Li Y-C, Li C-F, Chen L-B, et al. MicroRNA-766 targeting regulation of SOX6 expression promoted cell proliferation of human colorectal cancer. Onco Targets Ther. 2015;8:2981. doi:10.2147/OTT.S89459
52. Sand M, Skrygan M, Georgas D, et al. Microarray analysis of microRNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2012;68(3):119–126. doi:10.1016/j.jdermsci.2012.09.004
53. Li X, Shi Y, Yin Z, Xue X, Zhou B. An eight-miRNA signature as a potential biomarker for predicting survival in lung adenocarcinoma. J Transl Med. 2014;12(1):159. doi:10.1186/1479-5876-12-159
54. Haihai L, Xuelian L, Lu W, et al. MicroRNAs contribute to promyelocyte apoptosis in As2O3-treated APL cells. Cell Physiol Biochem. 2013;32(6):1818–1829. doi:10.1159/000356615
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.