Back to Journals » Cancer Management and Research » Volume 12
Long Noncoding RNA PCAT18 Upregulates SPRR3 to Promote Colorectal Cancer Progression by Binding to miR-759
Authors Yang D, Li R, Xia J, Li W, Ma L, Ye L, Xue H
Received 17 July 2020
Accepted for publication 1 October 2020
Published 9 November 2020 Volume 2020:12 Pages 11445—11452
DOI https://doi.org/10.2147/CMAR.S272652
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Daqing Yang,1 Rizeng Li,1 Jianfu Xia,1 Wencai Li,1 Lili Ma,2 Lechi Ye,3 Haibo Xue4
1Department of Colorectal Surgery, Wenzhou Central Hospital, Wenzhou 325000, People’s Republic of China; 2Department of Rheumatology, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, People’s Republic of China; 3Department of Colorectal and Anal Surgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, People’s Republic of China; 4The First Department of Gastroenterology, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, People’s Republic of China
Correspondence: Haibo Xue
The First Department of Gastroenterology, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, People’s Republic of China
Email [email protected]
Background: Long noncoding RNAs (lncRNAs) play essential functions in the development of several cancers, including colorectal cancer (CRC). Nevertheless, how PCAT18 regulates CRC tumorigenesis remains unclear. In this research, we aimed to investigate the roles of PCAT18 in CRC.
Materials and Methods: qRT-PCR and Western blot were used to analyze RNA and protein levels. CCK8, colony formation, transwell and wound healing assays were utilized to analyze proliferation, migration and invasion. Luciferase reporter assay was used to analyze RNA interactions.
Results: PCAT18 was found to be highly expressed in CRC tissues and cells. PCAT18 level was positively correlated with lymph node metastasis and TNM stage. Functionally, PCAT18 silencing induced impairment of CRC proliferation, migration and invasion. Besides, PCAT18 was identified to inhibit miR-759. PCAT18 promotes SPRR3 expression through binding to miR-759. Furthermore, miR-759 inhibitors or SPRR3 ectopic expression partially rescued the abilities of proliferation, migration and invasion in CRC cells transfected with sh-PCAT18.
Conclusion: Therefore, our study demonstrated that PCAT18 contributes to CRC progression through regulating miR-759/SPRR3 axis, which provides a new theoretical basis of explaining CRC tumorigenesis.
Keywords: long noncoding RNA, PCAT18, miR-759, SPRR3, colorectal cancer
Introduction
Colorectal cancer (CRC) is one of the most common cancers in the gastrointestinal tract and becomes a great health problem in the world.1 CRC progression is characterized with multiple stages and accumulation of genetic changes.2 Many pathways, such as Wnt signaling and PI3K/Akt signaling, areinvolved in CRC development.3 However, the molecular mechanism underlying CRC tumorigenesis still remains elusive. The prognosis of CRC patients is unsatisfactory and it is necessary to develop novel therapeutic methods.
Long noncoding RNA (lncRNA) is characterized by over 200 nucleotides in length and lacking protein-coding potential.4 Growing researches reveal that lncRNA plays multiple functions in several biological processes, including development, immune response and cancer.5,6 The significance of lncRNA in tumor has been gradually discovered. lncRNA may affect the proliferation, metastasis, survival or stem-ness of tumor cells.7 For example, lncRNA MAFG-AS1 initiates CRC growth and invasion through interacting with miR-147b to activate NDUFA4 expression.8 LncRNA TUC338 contributes to lung cancer cell proliferation and migration via promoting MAPK signaling.9 In addition, lncRNA FBXL19-AS1 contributes to the growth and migration of breast cancer through sponging miR-718.10 Therefore, there is an important requirement to define the correlation between lncRNA and cancer.
LncRNA PCAT18 has been shown to inhibit gastric cancer progression.11 Another study also confirms the inhibitory role of PCAT18 in gastric cancer.12 Nevertheless, the role of PCAT18 in CRC remains unclear. In this research, PCAT18 expression was found to be raised in CRC tissues and cells. PCAT18 upregulation predicted a poor prognosis. Moreover, PCAT18 knockdown suppressed the proliferation, migration and invasion of CRC cells. Mechanistically, PCAT18 was demonstrated to sponge miR-759 to upregulate SPRR3 expression. In conclusion, this study reveals the critical role of PCAT18/miR-759/SPRR3 axis in initiating CRC progression.
Materials and Methods
Patients and Tissues
Forty-three human CRC tissues and corresponding adjacent normal tissues were collected from the First Affiliated Hospital of Wenzhou Medical University. This study was approved by the Institutional Review Board of the First Affiliated Hospital of Wenzhou Medical University. Written informed consent was collected from all participants. All patients were not treated with chemotherapy or radiotherapy before surgery.
Cell Culture and Transfection
CRC cell lines and normal colon epithelial cell line FHC were bought from ATCC. Cell lines were cultured with DMEM (Gibco, Grand Island, NY, USA) containing 10% FBS (Gibco). Scrambled shRNA (sh-NC), PCAT18 shRNAs (sh-PCAT18), miR-759 inhibitors, miR-759 mimics and negative controls were purchased from GenePharma (Shanghai, China). Lipofectamine 2000 (Invitrogen Co., Carlsbad, CA, USA) were utilized for cell transfection.
Dual-Luciferase Reporter Assay
The wild-type or mutant sequences of PCAT18 or SPRR3 containing the putative binding site with miR-759 were inserted into luciferase reporter vector psi-CHECK-2 (Promega, Madison, WI, USA). For luciferase reporter assay, luciferase reporter and miR-759 mimics were transfected into CRC cells for 24 h. Then, the luciferase reporter activity was measured using the dual-luciferase reporter assay system (Promega, Madison, WI, USA).
Quantitative Real-Time PCR Analysis
Total RNA was isolated from tissues or cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA was reversely transcribed into cDNA via a Reverse Transcription Kit (QIAGEN, Valencia, CA, USA). qPCR was analyzed through SYBR-Green-quantitative real-time PCR Master Mix kit (Toyobo Co., Osaka, Japan). The mirVanaTM qRT-PCR microRNA Detection Kit (Ambion Inc., Austin, TX, USA) was utilized to detect miRNA expression. miRNA expression was normalized to U6. lncRNA and mRNA expression was normalized to GAPDH. Fold change was determined through the 2−ΔΔCt method. Primer sequences were available if requested.
CCK8 and Colony Formation Assays
Cell proliferation was detected using CCK8 and colony formation assays. For CCK8 assay, cells were seeded into 96-well plates and cultured for indicated days. Then, 10 μL of CCK8 solution was added into each well and incubated for 2 h. The absorbance at 450 nm was determined using a microplate reader (Bio-Rad, Hercules, CA, USA). For colony formation assay, cells were seeded into 6-well plates. Then, cells were cultured for 14 days. Colonies were fixed with 10% formaldehyde and stained with 0.5% crystal violet. Colonies numbers were finally counted.
Migration and Invasion Assays
Cell migration and invasion were measured through Transwell assay according to a previous study.13 Cell number per group was from five random fields. And three independent experiments were performed.
Statistical Analysis
SPSS software and Graphpad Prism software were used to conduct statistical analyses. Results were analyzed using t-test or a one-way analysis of variance and expressed as the mean ± standard deviation (SD). Survival curve was generated using the Kaplan–Meier method, and the differences were analyzed using the Log rank test. P < 0.05 was considered statistically significant.
Results
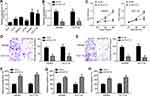
PCAT18 Was Upregulated in CRC Tissues
To explore the potential role of PCAT18, we firstly evaluated its expression patterns in CRC tissues. Results showed that PCAT18 was upregulated in CRC tissues compared to normal adjacent tissues (Figure 1A). Moreover, we noticed that PCAT18 level was higher in CRC tissues with lymph node metastasis (Figure 1B). And PCAT18 level was raised in tumor tissues of advanced stages (Figure 1C). Then, CRC tissues were divided into two groups based on PCAT18 level. We found that PCAT18 level was negatively correlated with patients’ prognosis (Figure 1D).
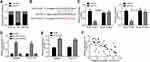
PCAT18 Knockdown Suppressed CRC Progression
Afterwards, the level of PCAT18 in CRC cell lines was analyzed. PCAT18 was highly expressed in most CRC cell lines (Figure 2A). Because PCAT18 expression levels were the highest in HCT116 and SW480 cells among all measured cell lines, we selected these two cells for further investigation (Figure 2A). We then knocked down PCAT18 in SW480 and HCT116 cells (Figure 2B). CCK8 assay indicated that PCAT18 knockdown suppressed proliferation rate of SW480 and HCT116 cells (Figure 2C). Besides, PCAT18 knockdown inhibited migration and invasion of CRC cells (Figure 2D and E). To further confirm the role of PCAT18, we overexpressed it. CCK8 and Transwell assays indicated that PCAT18 upregulation promoted proliferation, migration and invasion (Figure 2F–H). Thus, PCAT18 is an oncogenic lncRNA in CRC.
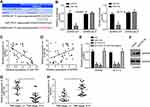
PCAT18 Interacted with miR-759
It is noticed that PCAT18 was mainly expressed in the cytoplasm of SW480 cells (Figure 3A), suggesting PCAT18 may be the sponge for some miRNAs. Through bioinformatics using miRDB, we identified miR-759 as the target of PCAT18 and constructed luciferase reporter vectors (Figure 3B). Luciferase reporter result supported that miR-759 mimics only suppressed the activity of WT-PCAT18 reporter (Figure 3C). RNA pulldown assay also showed that biotin-labeled miR-759 enriched PCAT18 (Figure 3D), demonstrating that PCAT18 was the sponge for miR-759. We also found that PCAT18 knockdown led to upregulation of miR-759 (Figure 3E). Additionally, PCAT18 level was negatively correlated with miR-759 in CRC tissues (Figure 3F).
MiR-759 Targeted SPRR3 Directly
Next, we used TargetScan to analyze the potential targets of miR-759. We identified SPRR3 as the most potential target and also constructed luciferase reporter vectors (Figure 4A). Similarly, miR-759 only suppressed the luciferase activity of WT-SPRR3 reporter (Figure 4B). Furthermore, miR-759 was negatively correlated with SPRR3 level in CRC tissues (Figure 4C) while SPRR3 expression was positively correlated with PCAT18 (Figure 4D). We also noticed that miR-759 mimics or PCAT18 knockdown suppressed SPRR3 expression (Figure 4E). Moreover, miR-759 inhibitors rescued the expression of SPRR3 in sh-PCAT18 transfected cells (Figure 4E). Consistently, SPRR3 expression was upregulated by PCAT18 overexpression (Figure 4F). Notably, we noticed that either miR-759 or SPRR3 was associated with tumor stages (Figure 4G and H).
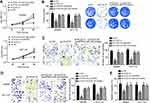
PCAT18 Promoted CRC Progression by Regulating miR-759/SPRR3 Axis
To determine whether PCAT18 regulates CRC progression via miR-759/SPRR3 axis, we performed CCK8, colony formation, Transwell and wound healing assays. Results indicated that PCAT18 knockdown repressed the proliferation, colony formation, migration and invasion of CRC cells (Figure 5A–E). However, miR-759 inhibition or SPRR3 overexpression restored the proliferation, colony formation, migration and invasion of CRC cells transfected with sh-PCAT18 (Figure 5A–E). Summarily, PCAT18 promotes CRC development through regulating miR-759/SPRR3 signaling.
Discussion
Determining the potential functions of lncRNAs in CRC is important and urgent. In this study, we explored the roles of PCAT18 in CRC progression. We found that PCAT18 was upregulated in CRC tissues. Its level was positively correlated with tumor metastasis and advanced stages. Moreover, PCAT18 upregulation indicated poor prognosis. PCAT18 knockdown suppressed CRC cell proliferation, migration and invasion. Therefore, our work demonstrated that PCAT18 was an important novel oncogene in CRC.
Increasing evidences have acknowledged the importance of lncRNAs in human cancers, such as CRC and cervical cancer.14,15 Notably, several lncRNAs have been proven to participate in CRC development. For example, lncRNA NEAT1 initiates CRC development through interacting with DDX5 to activate Wnt pathway.16 LncRNA SNHG7 promotes CRC growth and invasion through sponging miR-216b to upregulate GALNT1 expression.13 LncRNA DUXAP8 contributes to CRC cell growth and metastasis through regulating EZH2/LSD1 complex.17 Previously, PCAT18 was identified as a tumor suppressor in gastric cancer.12,18 However, the role of PCAT18 in other cancers, including CRC, remains fully unclear. In this work, we showed that PCAT18 was highly expressed in CRC tissues. Moreover, PCAT18 level was positively correlated with metastasis and advanced stage. Through functional experiments, we demonstrated that PCAT18 knockdown suppressed cell proliferation, migration and invasion in CRC. Therefore, our findings discovered that PCAT18 was an oncogenic gene in CRC.
LncRNAs have been demonstrated to serve as sponges of miRNAs.13 The lncRNA/miRNA network plays essential roles in regulating tumorigenesis.8 For instance, LncRNA-CDC6 sponges miR-215 to upregulate CDC6 and induces breast cancer development.19 LncRNA CDKN2BAS sponges miR-153 to increase ARHGAP18 level and causes liver cancer progression.20 Additionally, lncRNA XIST suppresses breast cancer growth and metastasis through sponging miR-155.21 PCAT18 has been found to sponge miR-107 and miR-135b in gastric cancer.11,12 We also found that PCAT18 was expressed in the cytoplasm of CRC cells, suggesting it may be also a miRNA sponge. Through bioinformatics prediction, we identified miR-759 as the target. Then, luciferase reporter assay and RNA pulldown assay demonstrated their interaction. miR-759 was found to affect human cutaneous melanoma cells.22 Its role in CRC is unknown. In our study, we showed that miR-759 was suppressed by PCAT18. And PCAT18 level was negatively correlated with miR-759 in CRC tissues. Moreover, functional assays suggested that miR-759 suppressed CRC development. Thus, our data revealed miR-759 is a new tumor suppressor in CRC.
Then, the downstream target of miR-759 was predicted through bioinformatics analysis. We identified SPRR3 and demonstrated it was directly targeted by miR-759. Moreover, we also uncovered that SPRR3 expression was upregulated by PCAT18 through inhibiting miR-759 in CRC. SPRR3 is involved in several cancers. A study reveals that SPRR3 regulates glioma cell proliferation and migration.23 Another work finds that SPRR3 may be a biomarker for esophageal squamous.24 Other studies also suggest that SPRR3 promotes the development of breast cancer, lung cancer and CRC.25–27 Consistently, our work also found that SPRR3 is an oncogene to promote CRC progression. However, our findings further illustrated the molecular mechanism underlying the regulation of SPRR3 expression by PCAT18/miR-759.
Conclusively, our findings demonstrated that PCAT18 promotes CRC progression through miR-759/SPRR3 axis, implying that PCAT18 may be a novel therapeutic target for CRC treatment. However, there are some limitations in our work. The downstream molecular mechanisms of SPRR3 require further investigation. The association between PCAT18 expression and clinicopathological features is also needed to define in the future. Additionally, animal experiments will further support our conclusion.
Acknowledgments
This study was supported by National Natural Science Foundation of China (81572291) and Wenzhou scientific research project (Y20190424).
Disclosure
The authors report no conflicts of interest for this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
2. Pritchard CC, Grady WM. Colorectal cancer molecular biology moves into clinical practice. Gut. 2011;60(1):116–129. doi:10.1136/gut.2009.206250
3. Walther A, Johnstone E, Swanton C, et al. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9(7):489–499. doi:10.1038/nrc2645
4. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi:10.1016/j.cell.2013.02.012
5. Liu B, Ye B, Yang L, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18(5):499–508. doi:10.1038/ni.3712
6. Wu Y, Yang X, Chen Z, et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18:87.
7. Ma S, Yang D, Liu Y, et al. LncRNA BANCR promotes tumorigenesis and enhances adriamycin resistance in colorectal cancer. Aging (Albany NY). 2018;10:2062–2078.
8. Cui S, Yang X, Zhang L, Zhao Y, Yan W. LncRNA MAFG-AS1 promotes the progression of colorectal cancer by sponging miR-147b and activation of NDUFA4. Biochem Biophys Res Commun. 2018;506(1):251–258. doi:10.1016/j.bbrc.2018.10.112
9. Zhang YX, Yuan J, Gao ZM, Zhang ZG. LncRNA TUC338 promotes invasion of lung cancer by activating MAPK pathway. Eur Rev Med Pharmacol Sci. 2018;22:443–449.
10. Ding Z, Ye P, Yang X, Cai H. LncRNA FBXL19-AS1 promotes breast cancer cells proliferation and invasion via acting as a molecular sponge to miR-718. Biosci Rep. 2019;39.
11. Zhang XZ, Mao H-L, Zhang S-J, et al. lncRNA PCAT18 inhibits proliferation, migration and invasion of gastric cancer cells through miR-135b suppression to promote CLDN11 expression. Life Sci. 2020;249:117478. doi:10.1016/j.lfs.2020.117478
12. Chen P, Zhao X, Wang H, et al. The down-regulation of lncRNA PCAT18 promotes the progression of gastric cancer via MiR-107/PTEN/PI3K/AKT signaling pathway. Onco Targets Ther. 2019;12:11017–11031. doi:10.2147/OTT.S225235
13. Shan Y, Ma J, Pan Y, et al. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9(7):722. doi:10.1038/s41419-018-0759-7
14. Tang J, Yan T, Bao Y, et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat Commun. 2019;10(1):3499. doi:10.1038/s41467-019-11447-8
15. Liu Y, Yang Y, Li L, et al. LncRNA SNHG1 enhances cell proliferation, migration, and invasion in cervical cancer. Biochem Cell Biol. 2018;96:38–43.
16. Zhang M, Weng W, Zhang Q, et al. The lncRNA NEAT1 activates Wnt/beta-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol. 2018;11(1):113. doi:10.1186/s13045-018-0656-7
17. Gong A, Huang Z, Ge H, Cai Y, Yang C. The carcinogenic complex lncRNA DUXAP8/EZH2/LSD1 accelerates the proliferation, migration and invasion of colorectal cancer. J BUON. 2019;24:1830–1836.
18. Foroughi K, Amini M, Atashi A, et al. Tissue-specific down-regulation of the long non-coding RNAs PCAT18 and LINC01133 in gastric cancer development. Int J Mol Sci. 2018;19(12):3881. doi:10.3390/ijms19123881
19. Kong X, Duan Y, Sang Y, et al. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234(6):9105–9117. doi:10.1002/jcp.27587
20. Chen J, Huang X, Wang W, et al. LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR-153-5p/ARHGAP18 signaling axis. Aging (Albany NY). 2018;10:3371–3381.
21. Zheng R, Lin S, Guan L, et al. Long non-coding RNA XIST inhibited breast cancer cell growth, migration, and invasion via miR-155/CDX1 axis. Biochem Biophys Res Commun. 2018;498(4):1002–1008. doi:10.1016/j.bbrc.2018.03.104
22. Su Y, Yu Y, He D, et al. Targeting STAT3 restores BRAF inhibitor sensitivity through miR-759-3p in human cutaneous melanoma cells. Int J Clin Exp Pathol. 2018;11:2550–2560.
23. Liu Q, Zhang C, Ma G, Zhang Q. Expression of SPRR3 is associated with tumor cell proliferation and invasion in glioblastoma multiforme. Oncol Lett. 2014;7(2):427–432. doi:10.3892/ol.2013.1736
24. de a Simão T, Souza-Santos PT, de Oliveira DSL. Quantitative evaluation of SPRR3 expression in esophageal squamous cell carcinoma by qPCR and its potential use as a biomarker. Exp Mol Pathol. 2011;91(2):584–589. doi:10.1016/j.yexmp.2011.06.006
25. Kim JC, Yu JH, Cho YK, et al. Expression of SPRR3 is associated with tumor cell proliferation in less advanced stages of breast cancer. Breast Cancer Res Treat. 2012;133(3):909–916. doi:10.1007/s10549-011-1868-5
26. Cho D-H, Jo YK, Roh SA, et al. Upregulation of SPRR3 promotes colorectal tumorigenesis. Mol Med. 2010;16(7–8):271–277. doi:10.2119/molmed.2009.00187
27. Li Q, Wang Y, Hu R, Yang G. Dysregulation of SPRR3/miR-876-3p axis contributes to tumorigenesis in non-small-cell lung cancer. Onco Targets Ther. 2020;13:2411–2419. doi:10.2147/OTT.S245422
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.