Back to Journals » Cancer Management and Research » Volume 11
Long noncoding RNA MIR31HG abrogates the availability of tumor suppressor microRNA-361 for the growth of osteosarcoma
Authors Sun Y, Jia X, Wang M, Deng Y
Received 6 May 2019
Accepted for publication 4 July 2019
Published 30 August 2019 Volume 2019:11 Pages 8055—8064
DOI https://doi.org/10.2147/CMAR.S214569
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Yongjie Sun,1 Xinghao Jia,2 Mingxing Wang,1 Yiqi Deng1
1Department of Orthopedics, Shanxian Central Hospital Affiliated to Jining Medical University, Heze, Shandong 274300, People’s Republic of China; 2Department of Orthopedics, Shanxian Haijiya Hospital, Heze, Shandong 274300, People’s Republic of China
Correspondence: Yiqi Deng
Department of Orthopedics, Shanxian Central Hospital, No.1 Wenhua Road, Heze, Shandong 274300, People’s Republic of China
Tel + 86 1 596 588 6792
Email [email protected]
Purpose: Long noncoding RNA (LncRNA) containing microRNA host gene is an interesting type of LncRNA. MicroRNA-31 (miR-31)-host gene LncRNA (MIR31HG) have been recognized as an oncogene in many cancers, but not in osteosarcoma (OS). Interestingly, MIR31HG/miR-31 could not regulate each other’s expression in certain cancer, suggesting that the role of MIR31HG in cancer is independent of miR-31. We here investigated the function and potential mechanism of MIR31HG in OS.
Methods: OS tissues and adjacent non-tumor tissues (n=40) were collected to determine the expressions of MIR31HG by paired t-test. We here identified the miRNAs predicted to be bound to MIR31HG and investigated the impacts of MIR31HG on cell growth and metastasis of OS cells by CCK-8, flow cytometry, Transwell assay, Western blot, etc. in vitro and in vivo.
Results: MIR31HG was upregulated in OS tissues and OS cell lines. The patients with high expression of MIR31HG have high tumor stages and distant metastasis. Tumor suppressor miR-361, but not miR-31, was confirmed to be sponged directly by MIR31HG in OS cells and was down-regulated in OS cell lines. Knockdown of MIR31HG restored the expression of miR-361. Restoration of miR-361 level in Saos-2 and U2OS cells induced cell apoptosis and G1/S arrest, inhibited proliferation and migration, which was, however, abrogated by MIR31HG. Mechanistically, cell growth and metastasis-related target genes of MIR-361 including VEGF, FOXM1 and Twist were de-repressed in OS cells by MIR31HG overexpression, leading to upregulated BCL2, CCND1 and epithelial–mesenchymal transition (EMT) phenotype. Patients with high expression of MIR31HG also showed more VEGF, FOXM1 and Twist levels. Overexpression of MIR31HG in vivo also promoted tumor growth via inhibition of miR-361 signals and elevated the expression of VEGF, FOXM1 and Twist for tumor growth.
Conclusion: MIR31HG acts as an oncogene in OS for tumor progression via regulation of tumor suppressor miR-361 and its target genes.
Keywords: LncRNAs, MIR31HG, miR-361, osteosarcoma, proliferation
Introduction
Osteosarcoma (OS) is the most common type of primary malignant bone tumor occurring mostly in adolescents and young adults, which accounts for 3.4% of all childhood tumors.1 The 5-year survival rate has increased to 60–70% though the introduction of wide resection surgery combining chemoradiotherapy. However, genetic and biological complexity of OS render parts of patients resistant to cancer treatment and metastasis, leading to the unsatisfactory outcomes.2,3
On basis of the size and shapes, noncoding RNA (ncRNA) can be subdivided into small ncRNAs (<200 nt long), such as microRNAs (miRNAs), and long ncRNAs (LncRNAs), with a length >200 bp.4,5 However, high-throughput sequencing technology has been identified and demonstrated many ncRNAs play a vital role in the cellular multiple physiological and pathological processes, such as cell proliferation, apoptosis, differentiation and invasion.5 Dysregulation of LncRNAs have identified as oncogene (H19, Lnc-NEAT1, MEG3, Lnc-ATB, Lnc-CAF,etc.) or tumor suppressor (Lnc-NORAD, Lnc-TSLNC8, Lnc-CASC15-S, etc.) during the carcinogenesis of lung cancer, bladder cancer, oral cancer, breast cancer, neuroblastoma and OS.6–8 Currently, some LncRNAs have been reported to be involved in the regulation of OS progression with promising diagnostic or prognostic value.9–11 Interestingly, some LncRNAs harbor miRNAs within their exonic or intronic sequences and hence are referred to as miRNA-host gene LncRNAs (Lnc-MIRHGs).12 However, the role of Lnc-MIRHGs in OS is largely unknown.
MiRNA-31 host gene (MIR31HG), known as LOC554202, is located on chromosome 9 (9p21.3). LncRNA MIR31HG served as a poor prognosis factor and was upregulated in laryngeal squamous cell cancer, lung cancer and colorectal cancer via targeting HIF1A, p21, p16, miR-193, miR-214, etc. for tumor growth, metastasis or chemotherapy resistance.13–15 However, some studies also reported that MIR31HG was downregulated in esophageal squamous cell carcinoma, hepatocellular carcinoma and bladder cancer.16–18 Thus, the expression of MIR31HG in cancer has not been consistent. Of note, although the expression of MIR31HG positively correlated miR-31 in some cancers, knockdown of MIR31HG showed no effects on the miR-31 levels and indicated that MIR31HG exerts its tumor-regulatory role is independent of miR-31.14,19 Currently, the expression of MIR31HG and its function in OS remained to be elucidated.
In this study, we investigated the clinical significance and function of MIR31HG in OS. The potential miRNA interacting with MIR31HG was predicted by bioinformatics method. The roles of MIR31HG/miRNA in cell growth or migration of OS were determined by knockdown or expression in vitro and in vivo.
Materials and methods
OS samples collection
All methods used for this study were approved by the Ethics Committee of Shanxian Central Hospital, and this study was conducted in accordance with the Declaration of Helsinki. A total of 40 OS tissues and corresponding relative normal tissues were obtained in this study from primary OS patients underwent complete resection surgery at Shanxian Central Hospital between 2015 and 2019. Written informed consent was obtained from all of the patients. All patients’ slides were reviewed to confirm the diagnosis by the same pathologist. All tissues were immediately snapped frozen in liquid nitrogen and stored at −80°C. There were 19 males (47.5%) and 21 females (52.5%) with age ranging from 8 to 30 years (average 17.5 years). Tumor localization at diagnosis in tibia/femur 25 cases (62.5%) and in other sites 15 cases (37.5%). Fourteen patients (35%) were found to have distant metastasis.
RNA extraction and Rt-qPCR
Total RNAs were isolated from tissues or cell lines using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The purity of the solution was ideal when A260/A280 ranged from 1.8 to 2.1. Afterward, 1 μg total RNA of each group was reversely transcribed into complementary DNAs (cDNAs) using the PrimeScript RT reagent Kit (Takara, Dalian China). qRT-PCR experiments were completed using the SYBR Green Kit (Takara). The qRT-PCR results of gene MIR31HG, vegf, foxm1 and twist were normalized to GAPDH and the gene of miR-31, miR-361 were normalized to U6 and expression fold change was calculated according to the 2−ΔΔCt method. The primers for MIR31HG, FP: TTCTGTCCTCCTACTCGGACCC, RP: CCTCCAGAGTTTGGTTTTGTGTC; miR-361, FP: TCCCCCAGGTGTGATTCTGAT; GAPDH, FP:TGACAACTTTGGTATCGTGGAAGG-3, RP:GCAGGGATGATGTTCTGGAGAG; U6, FP: GCTTCGGCAGCACATATACTAAAAT, RP: CGCTTCACGAATTTGCGTGTCA.
Cell lines and culture
OS cell lines 143B, MG63, U2OS and Saos-2 cells were purchased from American Type Culture Collection and cultured in DMEM supplemented with 10% FBS (Gibco, Grand Island, NY, USA), 100 U/mL of penicillin and 100 mg/mL of streptomycin (Invitrogen). Cultures were maintained at 37°C in a humidified CO2 (5%) atmosphere. Human osteoblastic cell line hFOB1.19 cells are cultured in Ham’s F12/DMEM supplemented with 10% FBS, 100 U/mL penicillin and 100 mg/mL streptomycin. Cultures were maintained at 33.5°C in a humidified CO2 (5%) atmosphere.
Plasmid construction, siRNA and transfection
MIR31HG full length (pcDNA3.1-MIR31HG) plasmid and controls (pcDNA3.1), miR-361 mimics and negative controls were synthesized by GenePharma (Shanghai, China). Two siRNAs targeting MIR31HG were synthesized and generated by GenePharma (Shanghai, China). The sequence for MIR31HG siRNA was: siRNA‑1, 5ʹ‑GCAAAGAAGUCCGAGGC‑3ʹ; siRNA‑2, 5ʹ‑GAGAAGAAAGAAGUCACC‑3ʹ. For transfection, U2OS and Saos-2 cells were cultured up to 60% confluency and transfected as indicated using lipofectamine 3000 (Invitrogen, USA) by incubating with OptiMem media for 4 hrs according to the manufacturer’s instructions.
Luciferase reporter assay
The MIR31HG cDNA fragment containing the predicted miR-361 binding site (Wt) and the matching mutant (Mut) was constructed into a psiCHECK2 vector. Renilla luciferase acted as control reporter (Promega, Madison, WI, USA). MiR-361 mimics or negative controls (100 nM) were transfected using Lipofectamine 2000. Luciferase activity was measured via Promega Dual-Luciferases Reporter Assay kit (Promega E1980) according to the manufacturer’s protocol after transfection. Relative Renilla luciferase activity was normalized to firefly luciferase activity.
CCK-8
The U2OS and Saos-2 cells were overexpressed miR-361 and/or MIR31HG, negative control, and then the cell proliferation was determined by using the Cell Counting Kit-8 (CCK-8, Beyotime, China) Kit. In brief, the CCK-8 solution was added into the wells and incubated for 2 hrs at 37°C. The absorbance at 450 nm was determined by a microplate reader (Bio-Rad, Carlsbad, CA) at the wavelength of 450 nm. All reactions were repeated in triplicate.
Cell apoptosis and cycle analysis
The U2OS and Saos-2 cells were overexpressed miR-361 and/or MIR31HG, negative control, and then were harvested and re-suspended in binding buffer containing Annexin V-FITC and PI according to the manufacturer’s instructions. The samples were analyzed by flow cytometry (BD Biosciences, USA). The percentages of apoptotic cells from each group were compared.
The cells for cell cycle analysis were fixed with 70% ice-cold ethanol for 48 hrs at 4°C and were rinsed with cold PBS followed by incubation with PBS containing 10 mg/mL PI and 0.5 mg/mL RNase A for 30 mins at 37°C. The DNA content of labeled cells was acquired using FACS cytometry assay (BD Biosciences, USA). Experiments were performed three times.
Transwell assay
A total of 2x105 transfected cells were re-suspended in DMEM and the cell suspension was seeded into the upper chamber containing a filtration membrane with a pore size of 8 μm. Then, 600 μL of medium containing 10% FBS was added into the lower chambers. After incubation for 24 hrs at 37°C, the cells remaining on the upper surface of the membrane were rubbed away with a sterile cotton swab and the migrated cells were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet (Beyotime, Nantong, China). Thereafter, the results of cell migration were observed, photographed and cells were enumerated in 5 random fields with an optical microscope.
Western blot
Ice-cold RIPA lysis buffer (Beyotime, Shanghai, China) including protease inhibitors was used to extract the total proteins from the cells, and protein concentrations were determined by using the Bradford assay (Bio-Rad, PA, USA). The proteins were subjected to SDS-PAGE on a 12% polyacrylamide gel and then electrophoretically transferred to polyvinylidene fluoride (PVDF) membranes (Merck Millipore, MA, USA). After blocking in 5% nonfat dry milk in Tris-buffered saline (TBS), the membranes were incubated with antibody against VEGF, FOXM1, Twist, BCL2, CCND1, E/N-cadherin and GAPDH (Abcam (USA), Cell Signaling Tech (Denver, MA)); then, the membranes were immunoblotted with horseradish peroxidase-conjugated secondary antibody (1:10,000 dilution) for 1 hr, before being detected by ECL Plus Western Blotting Substrate (Thermo Scientific, Shanghai, China).
Tumor model
Saos-2 cells (2×106) stably expressing MIR31HG, miR-361 mimics or empty vector by lentivirus infection were subcutaneously injected into male NOD/SCID mice (Shanghai Laboratory Animal Research Center, Shanghai, China). The tumors in each group were the same at the beginning of experiment and grew with time. The lentivirus was purchased from Genepharma (Shanghai, China). The volumes of tumors in each group were determined at day 7 and grew with time, and the difference between three groups appeared at day 10 although the tumor was small. The mice (n=5/group) were sacrificed at 28 days after tumor xenografting. Tumor volume was measured every 3 days and tumor tissues were obtained for qRT-PCR and Western blot analysis. The volume was calculated by the formula: length×width2×0.5. Animal studies were conducted in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and were approved by the animal ethics committee of Shanxian Central Hospital.
Immunohistochemistry
Fresh tumor tissues from nude mice were fixed with 4% paraformaldehyde and then embedded in paraffin to obtain tissue sections, which were then incubated at 4°C overnight with primary antibodies against CD31 (1:500; Abcam) and Ki-67 (1:400; Abcam). The sections were incubated with Envision System HRP-labeled polymer anti-rabbit (Dako K4003) or anti-mouse secondary antibodies (Dako K4001).
Statistical analysis
The Statistical Package for Social Sciences version 16.0 (SPSS 16.0, SPSS Inc., Chicago, IL, USA) and the Prism statistical software package (Version 5.0, Graphpad Software Inc.) were used in this study. We have conducted paired T-test to compare the expressions of MIR31HG and miR-361 the non-tumor and tumor groups. Unpaired t-tests or Mann–Whitney U tests were used to compare the two groups, and multiple group comparisons were analyzed with one-way ANOVA. P<0.05 was considered statistically significant. All experiments were performed at least three times.
Results
Overexpressed MIR31HG is associated with poor clinical outcomes of OS patients
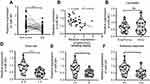
Considering the inconsistent expression trends of MIR31HG in different cancers, we here collected OS tissues and its corresponding non-tumor tissues (n=40) to determine the expression of MIR31HG during the carcinogenesis of OS. The results showed that MIR31HG was upregulated in OS tissues when compared to non-tumor tissues (Figure 1A). We further found that MIR31HG level in tumor tissues positively correlated with patients’ tumor size, Enneking stage and distant metastasis (Figure 1B–E), indicating that MIR31HG might participate into the progression of OS.
MIR31HG could directly interact with miR-361 in OS cell lines
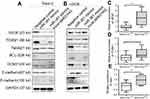
LncRNAs could sponge miRNAs to regulate the target genes of miRNAs during tumor initiation, progression. Since the function of MIR31HG is reported to be independent of miR-31 in pancreatic ductal adenocarcinoma,19 we determined the relationship in OS. We first confirm the increased expression of MIR31HG in OS cell lines 143B, MG63, U2OS and Saos-2 cells when compared to that in human osteoblastic cell line hFOB1.19 cells (Figure 2A). Knockdown of MIRHG31 in Saos-2 and U2OS cells has no effect on the miR-31 levels (Figure 2B), which was consistent with previous report.19
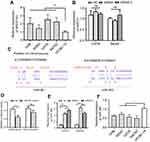 |
Figure 2 MIR31HG inhibits miR-361 level in OS cell lines. (A) The expression of MIR31HG was estimated in OS cell lines 143B, MG63, U2OS and Saos-2 cells and human osteoblastic cell line hFOB1.19 cells. (B) Knockdown of MIR31HG by two independent siRNA showed no effects on the miR-31 in U2OS and Saos-2 cells. (C) Bioinformatic analysis (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php) showed that MIR31HG harbor different binding sites of miR-361, blue letters showed the binding sites. (D) The Saos-2 cell line was transfected with luciferase reporters that contained MIR31HG with wild-type (WT) or mutated-type (MUT) miR-361 binding sites or miR-361 mimics, the luciferase activity was determined. (E) Knockdown of MIR31HG by two independent siRNA reduced the miR-31 level in U2OS and Saos-2 cells. (F) The expression of miR-361 was estimated in OS cell lines 143B, MG63, U2OS and Saos-2 cells and human osteoblastic cell line hFOB1.19 cells. *P<0.05, **P<0.01, data represent the means±SD. All experiments were performed at least three times.Abbreviations: OC, osteosarcoma; NC, normal control; NS, non-significant. |
We next predicted the miRNA that interact with MIR31HG via bioinformatics website online (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php), the miR-361 was the promising candidate miRNA to interact with MIG31HG (Figure 2C). Interestingly, miR-361 was found to be a tumor suppressor in many cancers. Luciferase reporters that contained MIR31HG with wild-type (WT) or mutated-type (MUT) miR-361 binding sites were conducted. The results showed that miR-361 mimics could significantly impair the luciferase activity of wild-type MIR31HG, but not the mutant-type in Saos-2 cell line (Figure 2D). Knockdown of MIR31HG significantly restored the miR-361 levels in two cell lines (Figure 2E). Indeed, the expression of miR-361 was also reduced in OS cell lines (Figure 2F). These data indicated the direct interaction between MIR31HG and miR-361 in OS.
MiR-361 is a tumor suppressor during the development of OS
Similarly, we determined the clinical significance of miR-361 in OS. The results showed that miR-361 was downregulated in OS tissues (Figure 3A), and the decreased expression of miR-361 was associated with high expression of MIR31HG (Figure 3B), Enneking stage and distant metastasis of OS patients, indicating its tumor-suppressive role in OS (Figure 3C–F).
MIR31HG/miR-361 signal regulates OS cell growth and migration in vitro
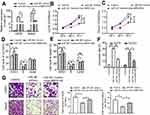
The function of MIR31HG/miR-361 signal was estimated, including cell vitality, apoptosis, cell cycle and cell migration. Saos-2 and U2OS cells transfected with pcDNA3.1-MIR31HG showed reduced expression of miR-361 (Figure 4A). Moreover, the results showed that overexpression of miR-361 mimics in Saos-2 and U2OS cells impaired the cell vitalities (Figure 4B and C) and the ability of migration (Figure 4G), induced G1/S arrest (Figure 4D and E) and cell apoptosis (Figure 4F). However, the anti-tumor effects of miR-361 were abrogated by overexpressed MIR31HG. These data demonstrated that MIR31HG could inhibit the availability of miR-361 for OS cell growth and migration.
MIR31HG regulates the target genes of miR-361 in OS patients
To investigate the mechanism of MIR31HG/miR-361 signal in OS progression, we speculated that the target genes of miR-361 were restored by overexpressed MIR31HG in OS cell lines. VEGF, FOXM1 and Twist were prominent target genes of MIR-361 for tumor growth, anti-apoptosis and metastasis.20–23 Overexpression of miR-361 reduced the expression of VEGF, FOXM1 and Twist in Saos-2 and U2OS cells, but MIR31HG could restore the expressions of these genes (Figure 5A and B) and upregulated anti-apoptosis BCL2 level and CCND1 level. Moreover, MIR31HG also promoted the EMT phenotype of Saos-2 and U2OS via miR-361/Twist signal.
The high and low expression of MIR31HG in OS tissues was determined by median. OS tissues with high expression of MIR31HG also have more expression of VEGF, FOXM1 and Twist, indicating that MIR31HG restored miR-361 signals in OS patients (Figure 5C-E).
MIR31HG/miR-361 signals participate in the tumor growth in vivo
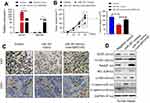
To further verify the function of MIR31HG/miR-361 signal in OS in vivo, Saos-2 cells stably expressing MIR31HG, miR-361 mimics or empty vector by lentivirus infection were subcutaneously injected into NOD/SCID mice (n=5/group) (Figure 6A). The results showed that mice overexpressed of miR-361 showed diminished tumor volume and delayed tumor growth (Figure 6B), but MIR31HG overexpression could inhibit the function of miR-361 and restore the tumor growth in parallel with increased expression of CD31 and Ki-67 (Figure 6C). The expressions of VEGF, FOXM1 and Twist were also restored by MIR31HG for tumor proliferation and EMT phenotype in tumor tissues (Figure 6D).
Discussion
We here firstly reported the diagnostic value and function of MIR31HG in OS, high expression of MIR31HG in OS tissues predicted advanced Enneking stage and distant metastasis of OS patients, and could promote OS progression via inhibition of tumor suppressor miR-361. This finding extended the family of LncRNA in the development of OS. Indeed, apart from our findings, there have been amounts of aberrantly expressed LncRNAs in tumor tissues or serum functioned as cancer biomarkers during the development of OS.10 The LncRNA Maternally expressed gene 3 (MEG3) was under-expressed in several human cancers, including non-small cell lung cancer, colorectal cancer and OS. LncRNA-TUG1 plasma-levels were found to correlate with disease status, with elevated levels indicating disease progression or recurrence.24 These researches indicated that LncRNAs could be promising diagnostic or prognostic value for OS patients, including the LncRNA MIRHG.
Since the feature that harboring miRNA host gene, LncRNA MIRHG also showed especial property differed from other LncRNAs. Lu et al found that the LncRNA MIR100HG and two embedded miRNAs, miR-100 and miR-125b, were overexpressed in the absence of known genetic events linked to cetuximab resistance via regulation of Wnt/β-catenin pathway.25 MIR503HG (miR-503 host gene) was highly expressed in anaplastic lymphoma kinase (ALK)-negative cell lines, which enhanced tumor cell growth by the induction of miRNA-503 (miR-503) and suppression of Smurf2, resulting in tumor cell growth.26 However, LncRNA MIRHG and its derived miRNA did not always reciprocally regulate their expressions and functions of each other. We here found that MIR31HG knockdown in two OS cell lines showed no effects on the expression of miR-31. This finding was consistent with other reports. Yang et al and Qin et al found that downregulation of MIR31HG had no effect on the expression of miR-31 in lung adenocarcinoma cells14 and pancreatic ductal adenocarcinoma specimens,19 although there was a significant positive correlation between MIR31HG and miR-31 in pancreatic ductal adenocarcinoma and colorectal cancer.27 These results implicated that MIR31HG exerts its function in miR-31-independent manner in cancer.
Some studies have focused on this question. Yan et al found that MIR31HG inhibited hepatocellular carcinoma proliferation and metastasis by sponging miRNA-575 to modulate ST7L expression.17 MIR31HG also act as an endogenous “sponge” by competing for miR-193b binding to regulate the miRNA targets for tumor cell growth.19 In addition, miR-214 was found to be inhibited by MIR31HG and overexpression of MIR31HG effectively reversed miR-214-induced inhibition of lung cancer progression.28 In this study, we identified that tumor suppressor miR-361 interacted with MIR31HG and was inhibited by overexpressed MIR31HG in OS cells. MIR31HG promoted cell proliferation and migration via regulating miR-361 signals, including upregulation of oncogene VEGF, FOXM1 and Twist. Of note, few studies have also determined miRNA-independent roles of LncRNA miRHGs. For example, LncRNA MIR100HG promoted cell proliferation in triple-negative breast cancer through triplex formation with p27 loci.29 All of these studies indicate miRNA-dependent or independent roles of MIR31HG in various key biological processes.
Conclusion
In sum, we identified an LncRNA MIR31HG/miR-361 signature with diagnostic value in OS and found that MIR31HG functioned as an oncogene to regulated miR-361 and its target genes VEGF, FOXM1 and Twist for tumor growth.
Acknowledgments
The research is supported by the Supporting Fund for Teachers’ research of Jining Medical University (No: JYFC2018FKJ090 for Yiqi Deng).
Author contributions
All authors contributed toward data analysis, drafting and revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92. doi:10.1007/978-3-319-07323-1_4
2. Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47(1):283–292. doi:10.1016/j.ocl.2015.08.022
3. Huang X, Zhao J, Bai J, et al. Risk and clinicopathological features of osteosarcoma metastasis to the lung: a population-based study. J Bone Oncol. 2019;16:100230. doi:10.1016/j.jbo.2019.100230
4. Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474(24):4219–4251. doi:10.1042/BCJ20170079
5. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi:10.1038/nrc.2017.99
6. Zheng Y, Liu L, Shukla GC. A comprehensive review of web-based non-coding RNA resources for cancer research. Cancer Lett. 2017;407:1–8. doi:10.1016/j.canlet.2017.08.015
7. Patel JS, Hu M, Sinha G, et al. Non-coding RNA as mediators in microenvironment-breast cancer cell communication. Cancer Lett. 2016;380(1):289–295. doi:10.1016/j.canlet.2015.11.016
8. Kita Y, Yonemori K, Osako Y, et al. Noncoding RNA and colorectal cancer: its epigenetic role. J Hum Genet. 2017;62(1):41–47. doi:10.1038/jhg.2016.66
9. Yang Z, Li X, Yang Y, He Z, Qu X, Zhang Y. Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death Dis. 2016;7(9):e2389. doi:10.1038/cddis.2016.272
10. Chen R, Wang G, Zheng Y, Hua Y, Cai Z. Long non-coding RNAs in osteosarcoma. Oncotarget. 2017;8(12):20462–20475. doi:10.18632/oncotarget.14726
11. Huang J, Deng G, Liu T, Chen W, Zhou Y. Long noncoding RNA PCAT-1 acts as an oncogene in osteosarcoma by reducing p21 levels. Biochem Biophys Res Commun. 2018;495(4):2622–2629. doi:10.1016/j.bbrc.2017.12.157
12. Sun Q, Tripathi V, Yoon JH, et al. MIR100 host gene-encoded lncRNAs regulate cell cycle by modulating the interaction between HuR and its target mRNAs. Nucleic Acids Res. 2018;46(19):10405–10416. doi:10.1093/nar/gky696
13. Wang R, Ma Z, Feng L, et al. LncRNA MIR31HG targets HIF1A and P21 to facilitate head and neck cancer cell proliferation and tumorigenesis by promoting cell-cycle progression. Mol Cancer. 2018;17(1):162. doi:10.1186/s12943-018-0916-8
14. Qin J, Ning H, Zhou Y, Hu Y, Yang L, Huang R. LncRNA MIR31HG overexpression serves as poor prognostic biomarker and promotes cells proliferation in lung adenocarcinoma. Biomed Pharmacother. 2018;99:363–368. doi:10.1016/j.biopha.2018.01.037
15. Montes M, Nielsen MM, Maglieri G, et al. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat Commun. 2015;6:6967. doi:10.1038/ncomms7967
16. Ren ZP, Chu XY, Xue ZQ, et al. Down-regulation of lncRNA MIR31HG correlated with aggressive clinicopathological features and unfavorable prognosis in esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2017;21(17):3866–3870.
17. Yan S, Tang Z, Chen K, et al. Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J Exp Clin Cancer Res. 2018;37(1):214. doi:10.1186/s13046-018-0853-9
18. He A, Chen Z, Mei H, Liu Y. Decreased expression of LncRNA MIR31HG in human bladder cancer. Cancer Biomarker. 2016;17(2):231–236. doi:10.3233/CBM-160635
19. Yang H, Liu P, Zhang J, et al. Long noncoding RNA MIR31HG exhibits oncogenic property in pancreatic ductal adenocarcinoma and is negatively regulated by miR-193b. Oncogene. 2016;35(28):3647–3657. doi:10.1038/onc.2015.430
20. Wang HW, Lo HH, Chiu YL, et al. Dysregulated miR-361-5p/VEGF axis in the plasma and endothelial progenitor cells of patients with coronary artery disease. PLoS One. 2014;9(5):e98070. doi:10.1371/journal.pone.0098070
21. Cui W, Li Y, Xu K, et al. miR-361-5p inhibits hepatocellular carcinoma cell proliferation and invasion by targeting VEGFA. Biochem Biophys Res Commun. 2016;479(4):901–906. doi:10.1016/j.bbrc.2016.09.076
22. Gao F, Feng J, Yao H, Li Y, Xi J, Yang J. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):776–782. doi:10.1080/21691401.2019.1577883
23. Ihira K, Dong P, Xiong Y, et al. EZH2 inhibition suppresses endometrial cancer progression via miR-361/Twist axis. Oncotarget. 2017;8(8):13509–13520. doi:10.18632/oncotarget.14586
24. Smolle MA, Pichler M. The role of long non-coding RNAs in osteosarcoma. Non-coding RNA. 2018;4:1. doi:10.3390/ncrna4010007
25. Lu Y, Zhao X, Liu Q, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/beta-catenin signaling. Nat Med. 2017;23(11):1331–1341. doi:10.1038/nm.4424
26. Huang PS, Chung IH, Lin YH, Lin TK, Chen WJ, Lin KH. The long non-coding RNA MIR503HG enhances proliferation of human ALK-negative anaplastic large-cell lymphoma. Int J Mol Sci. 2018;19:5.
27. Eide PW, Eilertsen IA, Sveen A, Lothe RA. Long noncoding RNA MIR31HG is a bona fide prognostic marker with colorectal cancer cell-intrinsic properties. Int J Cancer. 2019;144(11):2843–2853. doi:10.1002/ijc.31998
28. Dandan W, Jianliang C, Haiyan H, Hang M, Xuedong L. Long noncoding RNA MIR31HG is activated by SP1 and promotes cell migration and invasion by sponging miR-214 in NSCLC. Gene. 2019;692:223–230. doi:10.1016/j.gene.2018.12.077
29. Wang S, Ke H, Zhang H, et al. LncRNA MIR100HG promotes cell proliferation in triple-negative breast cancer through triplex formation with p27 loci. Cell Death Dis. 2018;9(8):805. doi:10.1038/s41419-018-0869-2
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.