Back to Journals » Cancer Management and Research » Volume 12
Long Noncoding RNA LINC00839 Promotes the Malignant Progression of Osteosarcoma by Competitively Binding to MicroRNA-454-3p and Consequently Increasing c-Met Expression
Authors Zhang Y, Guo H, Ma L, Chen X, Chen G
Received 28 June 2020
Accepted for publication 15 August 2020
Published 24 September 2020 Volume 2020:12 Pages 8975—8987
DOI https://doi.org/10.2147/CMAR.S269774
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
This paper has been retracted.
Yang Zhang, 1 Hai Guo, 2 Li Ma, 3 Xiaoyong Chen, 1 Guangdong Chen 4
1Department of Orthopedics, Shenzhen University General Hospital, Shenzhen 518055, People’s Republic of China; 2Department of Anesthesiology, First Affiliated Hospital of Xinjiang Medical University, Urumqi, Xinjiang, 830000, People’s Republic of China; 3Department of Emergency, General Hospital of Xinjiang Military Command of Chinese People’s Liberation Army, Urumqi, Xinjiang, 830000, People’s Republic of China; 4Department of Orthopedics, Cangzhou Center Hospital, Cangzhou, Hebei 061014, People’s Republic of China
Correspondence: Yang Zhang Department of Orthopedics
Shenzhen University General Hospital, 1098 Xueyuan Road, Nanshan District, Shenzhen 518055, People’s Republic of China
Email [email protected]
Purpose: This study was conducted to determine the expression and prognostic relevance of long intergenic non-protein coding RNA 839 (LINC00839) in osteosarcoma (OS) and to explore the detailed roles of LINC00839 in regulating OS cell activities and the mechanisms responsible for its cancer-promoting activity in OS.
Methods: The expression of LINC00839 in OS tissues and cell lines was determined by quantitative reverse transcription–polymerase chain reaction. After LINC00839 knockdown, cell counting kit-8 assay, flow cytometric analysis, transwell migration and invasion assay, and in vivo tumor xenograft assay were used to detect its effects on cellular processes in OS. Bioinformatics analyses were conducted to predict the putative miRNAs that target LINC00839. RNA immunoprecipitation assay, luciferase reporter assay, Western blotting analysis, and rescue assays were conducted to establish a relationship among LINC00839, microRNA-454-3p (miR-454-3p), and cellular mesenchymal to epithelial transition factor (c-Met) in OS.
Results: LINC00839 was upregulated in OS tissues and cell lines. OS patients characterized with high LINC00839 expression exhibited shorter overall survival than patients with low LINC00839 expression. LINC00839 knockdown caused a significant reduction in OS cell proliferation, migration, and invasion in vitro. Furthermore, LINC00839 depletion inhibited OS tumor growth in vivo and induced apoptosis. Mechanistically, LINC00839 functions as a competitive endogenous RNA in OS by sponging miR-454-3p. c-Met was confirmed as a direct target gene for miR-454-3p in OS cells and was positively regulated by LINC00839 by competitively binding to miR-454-3p.
Conclusion: LINC00839 promoted the oncogenicity of OS by targeting the miR-454-3p/c-Met axis. The LINC00839/miR-454-3p/c-Met network may represent a potential target for OS therapy.
Keywords: long intergenic non-protein coding RNA 839, ceRNA, therapeutic target
Introduction
Osteosarcoma (OS) is the most frequently occurring primary musculoskeletal malignancy in children and adolescents.1 The morbidity of OS is approximately 4.4 individuals per million worldwide, and most of OS cases are diagnosed in developing countries and the least developed countries.2 The medullary ends of the long bones are a vulnerable site for OS, particularly in the distal femur and the proximal humerus.3 Improvements in diagnostic methods and treatment regimens have resulted in increased survival rates for OS patients over the last decades.4 Yet, the prognosis of OS remains dismal, especially for patients with distant metastasis or recurrent disease.5 Difficulties in treating this disease are primarily due to its characteristically destructive and highly metastatic potential.6 Additionally, the poor outcomes observed for OS are also closely related to our limited understanding of the mechanisms underlying OS oncogenesis and progression.7 Therefore, it is necessary to elucidate the molecular events associated with OS pathogenesis, which may facilitate the development of effective targets for cancer diagnosis, prognosis, and therapy.
Long noncoding RNAs (lncRNAs) belong to a group of noncoding RNA molecules that are over 200 nucleotides in length.8 LncRNAs do not encode proteins because they do not contain an open reading frame. Originally, lncRNAs were thought to represent “noise” from the gene transcription process.9 Instead, recent studies have indicated that lncRNAs participate in nearly all physiological and pathological processes and that these actions occur through direct or indirect control of protein expression.10,11 The aberrant expression of lncRNAs is prevalent in a number of human cancer types. With regard to OS, studies have indicated that many lncRNAs are differentially expressed in OS and may serve as prognostic biomarkers.12–14 The aberrantly expressed lncRNAs affect the oncogenicity of OS and play a role in tumor initiation or inhibition.15–17
MicroRNAs (miRNAs) refer to a group of short, noncoding RNAs of approximately 18–23 nucleotides in length.18 They are considered regulators of gene expression by inducing translation suppression or promoting mRNA degradation through direct binding to the complementary 3′-untranslated regions of their target genes in a base-pairing manner.19 In recent years, a competing endogenous RNA (ceRNA) theory was proposed by Leonardo Salmena, which has now become widely accepted.20 It was proposed that lncRNA possesses microRNA response elements that function as an miRNA sponge, which attenuate miRNA-mediated regulation of target genes.21 Therefore, the ceRNA network may represent a promising and effective target for OS prognosis, prevention, and therapy.
A number of lncRNAs have been identified and verified throughout the human genome;22 however, their specific roles in OS cells require further study. The expression and function of LINC00839 in OS has not been well addressed, which has prompted us to investigate whether LINC00839 contributes to OS progression. In this study, we initially measured the expression of LINC00839 in OS and determined its prognostic relevance. Then, a series of loss-of-function experiments were conducted to explore the role of LINC00839 in regulating OS cell processes. Furthermore, mechanistic studies were done to address the mechanisms underlying the oncogenic role of LINC00839 in OS cells.
Materials and Methods
Tissue Specimens
The collection and use of tissue samples were done with approval from the Ethics Committee of Shenzhen University General Hospital (2017–0804) and performed in accordance with the Declaration of Helsinki. Written informed consent was also provided by all participants or their guardians. OS tissues and corresponding adjacent normal tissues were collected from 47 OS patients at Shenzhen University General Hospital. All participants had not received preoperative radiotherapy, chemotherapy, or other anticancer therapies. Fresh tissues were collected, immediately frozen in liquid nitrogen, and stored in liquid nitrogen until use.
Cell Lines
The hFOB 1.19 normal osteoblast cell line was acquired from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China), and cultured in D-MEM/F-12 medium supplemented with 10% fetal bovine serum (FBS) and 0.3 mg/mL G418 (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Two OS cell lines, MG-63 (Shanghai Institute of Biochemistry and Cell Biology) and Saos-2 (Shanghai Institute of Biochemistry and Cell Biology), were maintained in MEM medium (Gibco; Thermo Fisher Scientific) and McCoy’s 5A medium (Gibco; Thermo Fisher Scientific), respectively, and both were supplemented with 10% FBS and 1% penicillin/streptomycin mixture (Gibco; Thermo Fisher Scientific). Two additional OS cell lines, HOS and U-2OS, were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). HOS cells were grown in ATCC-formulated Eagle’s Minimum Essential Medium (ATCC) containing 10% FBS, whereas 10% FBS-supplemented McCoy’s 5a Medium Modified (Gibco; Thermo Fisher Scientific) was used for culturing the U-2OS cell line. All cell lines were cultured in a humidified atmosphere at 37°C under 5% CO2.
Transient Transfection
The small interfering RNAs (siRNAs) targeting LINC00839 and corresponding control siRNA (si-NC) were obtained from Shanghai GenePharma Co., Ltd. (Pudong, Shanghai, China). The miR-454-3p mimic, negative control (NC) miRNA mimic (miR-NC), miR-454-3p inhibitor and NC inhibitor were constructed by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The c-Met overexpressing plasmid, pcDNA3.1-c-Met (pc-c-Met), and control pcDNA3.1 empty vector plasmid were also designed and synthesized by Shanghai GenePharma Co., Ltd. Cells were seeded into 6-well plates and Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for transient transfection of the siRNAs, miRNA mimic/inhibitor, or plasmids into OS cells.
Quantitative Reverse Transcription–Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated from tissues or cells using the RNAsimple Kit (TIANGEN; Beijing, China). A NanoDrop™ 2000 Spectrophotometer (Invitrogen; Thermo Fisher Scientific, Inc.) was used to assess the quality and concentration of RNA samples. Complementary DNA (cDNA) was obtained by performing reverse transcription using PrimeScript™ RT Master Mix (Perfect Real Time; Takara Bio, Inc., Tokyo, Japan). The expression of LINC00839 and c-Met was detected using TB Green® Premix Ex Taq™ II (Takara Bio, Inc.), which was conducted on an ABI 7900 system (Applied Biosystems, Foster City, CA, USA). GAPDH served as an internal reference for LINC00839 and c-Met expression.
To analyze miR-454-3p expression, total RNA was reverse transcribed into cDNA using miRcute Plus miRNA First-Strand cDNA Kit (TIANGEN). Then, miRcute Plus miRNA qPCR Kit (SYBR Green; TIANGEN) was used for qPCR. The expression of miR-454-3p was normalized to that of U6 small nuclear RNA. All data were analyzed using the 2−ΔΔCt method.
Subcellular Fractionation Location
Isolation of RNA from the cytoplasmic and nuclear fractions was done using the Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA). qRT-PCR was used to quantitate LINC00839, GAPDH and U6 in the RNA samples. GAPDH was used as the cytoplasmic internal reference and U6 was used as the nuclear RNA control.
Cell Counting Kit-8 (CCK-8) Assay
A cell suspension (100 μL) containing 2 × 103 cells was seeded into 96-well plates. Each group contained five replicates. CCK-8 assay was conducted every 24 h for 72 h. At each time point, 10 μL of CCK-8 solution (Dojindo Laboratories Co. Ltd., Kumamoto, Japan) was incubated with the transfected cells at 37°C with 5% CO2 for 2 h. The absorbance value of each well was measured at 450 nm using a microplate reader (Tecan Infinite M200 Micro Plate Reader; LabX, Switzerland).
Flow Cytometric Analysis
OS cells transfected with the aforementioned oligonucleotides or plasmid were incubated for 48 h in a humidified atmosphere at 37°C with 5% CO2. Cell apoptosis was analyzed with an annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (Biolegend, San Diego, CA, USA). Cells were lysed using ethylenediaminetetraacetic acid-free trypsin (Gibco) and collected by centrifugation, followed by resuspension in 100 μL of binding buffer, and staining with 10 μL of Annexin V-FITC and 5 μL of propidium iodide for 15 min in the dark. Finally, the ratio of apoptotic cells was analyzed by flow cytometry (FACScan, Becton Dickinson, Franklin, NJ).
Transwell Migration and Invasion Assay
For migration assays, transfected cells were trypsinized at 48 h post-transfection, rinsed with phosphate buffer solution, collected via centrifugation and resuspended in FBS-free medium. The upper chambers of transwell inserts (BD Biosciences) were loaded with 200 μL cell suspension containing 5 × 104 cells, while basal medium supplemented with 10% FBS was added to the lower chambers. After 24 h, the non-migrated cells were removed with a cotton swab, while the migrated cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Subsequent to extensive washing, the migrated cells were imaged with an inverted microscope (IX31; Olympus Corporation, Tokyo, Japan), after which six visual fields were randomly selected and the average number of migrated cells was calculated. Transwell invasion assay was conducted using the same experimental protocols as the migration assay, with the exception that the transwell inserts were precoated with Matrigel (BD Biosciences).
Tumor Xenograft in vivo Assay
Lentiviral particles stably expressing the short hairpin RNA (shRNA) specific for LINC00839 (sh-LINC00839) and negative control shRNA (sh-NC) were produced by Shanghai GenePharma Co., Ltd. HOS cells were injected with lentiviruses and incubated with puromycin to select cells stably transfected with sh-LINC00839 or sh-NC.
The Ethics Committee of Animal Experiments at Shenzhen University General Hospital (2019–0211) approved the animal protocols. The study was conducted in strict accordance with the NIH guidelines for the care and use of laboratory animals (NIH Publication No. 85–23 Rev.1985). Male 4-week-old BALB/c nude mice were acquired from the Guangdong Medical Laboratory Animal Center (Guangdong, China) and subcutaneously injected with 5 × 106 HOS cells with the stable LINC00839 knockdown construct. The same number of HOS cells stably expressing sh-NC was also injected into nude mice as the control group. Following injection, tumor width (a) and length (b) were monitored every 5 days and tumor volume was calculated according to the formula: volume (mm3) = (a2 × b)/2. Finally, the mice were euthanized at 30 days after inoculation and the tumor xenografts were excised. After weighing, total RNA and protein were extracted and used for molecular assays.
Bioinformatics Prediction
StarBase version 3.0 (http://starbase.sysu.edu.cn/) and DIANA tools – LncBase Experimental v2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2findex-experimental) were used to search for the miRNAs that may be targeted by LINC00839.
RNA Immunoprecipitation (RIP) Assay
RIP assay was conducted using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA). OS cells were lysed in complete RIP lysis buffer and the cell lysate was incubated with RIP buffer containing magnetic beads conjugated with an anti-Argonaute 2 (Ago2) antibody or normal mouse IgG antibody (Millipore, Bedford, MA, USA). Following an overnight incubation at 4°C, the magnetic beads were harvested and rinsed with wash buffer. The resulting immunoprecipitate complex was incubated with proteinase K to purify RNA. Finally, the relative enrichment of LINC00839 and miR-454-3p in the immunoprecipitated RNA was determined by qRT-PCR.
Luciferase Reporter Assay
The fragments of LINC00839 and c-Met that harbor the predicted miR-454-3p binding site were synthesized and subcloned into the dual-luciferase reporter vector pmirGLO (Promega Corporation, Madison, WI, USA) to generate the LINC00839-wild-type (LINC00839-wt) and c-Met-wt reporter plasmids. At the same time, the reporter plasmids, LINC00839-mutant (LINC00839-mut), and c-Met-mut were constructed by mutating the binding sequences in LINC00839 and c-Met, respectively.
When OS cells reached approximately 60–80% confluence, the wt or mut reporter plasmids were cotransfected with miR-454-3p mimic or miR-NC using Lipofectamine® 2000. At 48 h post-transfection, cells were harvested and assayed for the measurement of luciferase activity using a Dual-Luciferase Reporter Assay System (Promega). Renilla luciferase activity was used to normalize the measurements.
Western Blotting Analysis
RIPA protein lysis buffer (TIANGEN) and the BCA Protein Assay Kit (TIANGEN) were used to isolate total protein and detect protein concentration, respectively. Equal amounts of protein extracts were separated on 10% sodium dodecyl sulfate-polyacrylamide gels and then transferred to PVDF membranes (Millipore, Billerica, MA, USA). After blocking with 5% non-fat milk at room temperature for 2 h, the membranes were incubated with primary antibodies against c-Met (ab216574; Abcam, Cambridge, UK) or GAPDH (ab181603; Abcam) overnight at 4°C. Subsequently, the membranes were incubated with a horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G secondary antibody (ab150077; Abcam) at room temperature for 2 h, followed by detection with the Immobilon® ECL Ultra Western HRP Substrate (Millipore). GAPDH was used as a loading control.
Statistical Analysis
All results are presented as the mean ± standard deviation from three independent experiments. A Student’s t-test was utilized for comparison between two groups, whereas the differences among three groups or more were assessed using a one-way analysis of variance in conjunction with Tukey’s test. The overall survival curves were generated using the Kaplan–Meier method. The Log rank test was applied to compare the differences between the overall survival curves. The correlation analysis between genes in the OS tissues was performed using the Pearson correlation coefficient. P values <0.05 were considered statistically significant.
Results
Depletion of LINC00839 Attenuates Cell Proliferation, Migration, Invasion and Facilitates Cell Apoptosis in OS
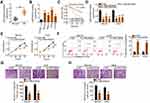
qRT-PCR was initially done to determine if LINC00839 was differentially expressed in OS. The results indicated that LINC00839 was significantly upregulated in OS tissues compared with that of adjacent normal tissues (Figure 1A). Expression of LINC00839 was also measured in a panel of OS cell lines (MG-63, Saos-2, HOS, and U-2OS) and normal hFOB 1.19 osteoblast cells as a control. The qRT-PCR data indicated that all four OS cell lines exhibited a high level of LINC00839 compared with that of hFOB 1.19 (Figure 1B). The clinical relevance of LINC00839 in OS was then examined. Using the median value of LINC00839 in OS tissues as the cutoff line, all enrolled OS patients were divided into either LINC00839 low or LINC00839 high expression groups. Kaplan–Meier analysis revealed that high LINC00839 expression had an adverse influence on the overall survival of patients with OS (Figure 1C; P = 0.039).
Having validated the aberrant upregulation of LINC00839 in OS, the functional role of this lncRNA in cancer progression was explored. Small interfering RNAs against LINC00839 (si-LINC00839) were transfected into MG-63 and HOS cells, which express relatively high LINC00839 levels among the four OS cell lines. qRT-PCR confirmed that the siRNAs silenced LINC00839 expression in MG-63 and HOS cells to varying degrees (Figure 1D). The si-LINC00839#1 exhibited the highest transfection efficiency and was therefore used in the subsequent functional experiments. The CCK-8 assay revealed that the proliferation of MG-63 and HOS cells was significantly hindered by si-LINC00839 transfection (Figure 1E). Furthermore, flow cytometric analysis demonstrated that the loss of LINC00839 caused a significant pro-apoptotic effect on MG-63 and HOS cells (Figure 1F). In addition, transwell migration and invasion assays were conducted to determine the effects of si-LINC00839 transfection on migration and invasion in OS cells. Knockdown of LINC00839 decreased the migratory (Figure 1G) and invasive (Figure 1H) capacities of MG-63 and HOS cells. In summary, these results suggest that LINC00839 exhibits a tumor-promoting function in regulating the progression of OS.
LINC00839 Acts as a Sponge for MiR-454-3p in OS Cells
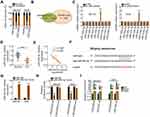
The functional roles of lncRNAs in human cancers are largely dependent on their location.21 To elucidate the mechanisms by which LINC00839 controls OS progression, the subcellular distribution of LINC00839 was analyzed by subcellular fractionation assay. The results corroborated that LINC00839 was mainly distributed in the cell cytoplasm of MG-63 and HOS cells (Figure 2A). Studies have reported that lncRNAs may function during oncogenesis and progression through ceRNA theory by working as a miRNA sponge.23 Bioinformatics tools were utilized to find the putative miRNAs containing a consensus binding sequence for LINC00839. StarBase 3.0 and DIANA tool – LncBase Experimental v2 predicted 33 and 465 miRNAs as potential targets, respectively (Figure 2B). In total, 27 miRNAs were simultaneously identified from the two data sets. MiR-106b-5p,24 miR-130a-3p,25 miR-130b-3p,26 miR-17-5p,27 miR-19a-3p,28 miR-19b-3p,29 miR-301a-3p,30 miR-4295,31 and miR-93-5p32 have been previously shown to be upregulated in OS and play oncogenic roles during cancer progression. In addition, no studies have reported an association of miR-1180-5p, miR-301b-3p, miR-3666, miR-519b-3p, miR-519c-3p, miR-520g-3p, miR-520h, miR-5590-3p, and miR-7114-3p with OS. Hence, these miRNAs were excluded from study, whereas nine miRNAs (miR-106a-5p, miR-20a-5p, miR-20b-5p, miR-223-3p, miR-338-3p, miR-454-3p, miR-519a-3p, miR-519d-3p, and miR-526b-3p) were selected for further analysis.
To test our hypothesis, LINC00839 depleted-MG-63 and HOS cells were subjected to qRT-PCR analysis for the detection of miRNA expression changes. Among these candidates, miR-454-3p was significantly increased in MG-63 and HOS cells by silencing LINC00839 expression, whereas the expression of the other miRNAs was unaffected in response to si-LINC00839 transfection (Figure 2C). Next, qRT-PCR analysis indicated that miR-454-3p was downregulated in OS tissues (Figure 2D), which was consistent with a previous study.33 Interestingly, the Pearson correlation analysis indicated an inverse correlation between LINC00839 and miR-454-3p in the OS tissues (Figure 2E; r = −0.7687, P < 0.0001). The wild-type and mutant binding sites of miR-454-3p within LINC00839 are presented in Figure 2F. A luciferase reporter assay was then used to determine whether LINC00839 could bind to miR-454-3p in OS cells. The efficiency of miR-454-3p mimic transfection was analyzed by qRT-PCR (Figure 2G). The results showed that resumption of miR-454-3p led to a significant reduction of luciferase activity in LINC00839-wt-transfected MG-63 and HOS cells; however, there was no significant effect on luciferase activity when both cells were cotransfected with LINC00839-mut and the miR-454-3p mimic (Figure 2H). Moreover, the RIP assay revealed that LINC00839 and miR-454-3p were preferentially enriched in Ago2-containing beads compared with the IgG group (Figure 2I), further indicating a direct binding relationship between miR-454-3p and LINC00839 in OS cells. Collectively, the results indicated that LINC00839 functions by sponging miR-454-3p in OS cells.
LINC00839 Upregulates c-Met in OS Cells by Sponging MiR-454-3p
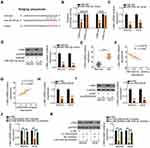
A previous study identified c-Met as a direct miR-454-3p target in OS cells.33 Therefore, we performed a series of experiments to confirm this result. The wild-type and mutant binding sites of miR-454-3p within the 3ʹ-UTR of c-Met are shown in Figure 3A. To validate this direct binding, luciferase reporter assays were performed. We found that cotransfection of miR-454-3p mimic and c-Met-wt significantly decreased luciferase activity in MG-63 and HOS cells when compared with cells cotransfected with c-Met-wt and miR-NC, whereas mutation of the binding sequences abolished the suppressive effect of miR-454-3p overexpression on luciferase activity (Figure 3B). Furthermore, the miR-454-3p mimic downregulated the expression of c-Met mRNA (Figure 3C) and protein (Figure 3D) in MG-63 and HOS cells. Also, c-Met was highly expressed in OS (Figure 3E) and inversely correlated with miR-454-3p expression (Figure 3F; r = −0.6976, P < 0.0001).
After establishing that LINC00839 worked by sponging miR-454-3p and that c-Met is a direct target of miR-454-3p, we hypothesized that LINC00839 regulates c-Met expression in OS cells by acting as a ceRNA. Initially, a positive correlation between LINC00839 and c-Met mRNA was observed in OS tissues (Figure 3G; r = 0.7216, P < 0.0001), as evidenced by Pearson correlation analysis. qRT-PCR and Western blotting analysis were used to measure c-Met expression in MG-63 and HOS cells after LINC00839 depletion. As expected, the levels of c-Met mRNA (Figure 3H) and protein (Figure 3I) were reduced in MG-63 and HOS cells following transfection with si-LINC00839, while introducing the miR-454-3p inhibitor almost restored c-Met expression levels (Figure 3J and K) that were suppressed by si-LINC00839. Altogether, LINC00839 worked as a ceRNA to competitively interact with miR-454-3p in OS cells and consequently increase c-Met expression.
Increasing the MiR-454-3p/c-Met Axis Output Abolishes the Regulatory Actions of LINC00839 Knockdown in OS Cells
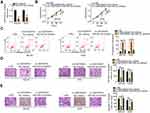
As miR-454-3p/c-Met has been determined to be modulated by LINC00839, rescue experiments were designed and conducted to confirm whether the miR-454-3p/c-Met axis is required for LINC00839-induced activities in OS cells. The miR-454-3p inhibitor was used and its silencing efficiency in MG-63 and HOS cells was verified by qRT-PCR (Figure 4A). Then, the miR-454-3p inhibitor or NC inhibitor together with si-LINC00839 were cotransfected into MG-63 and HOS cells. CCK-8 assay indicated that downregulation of miR-454-3p rescued the proliferative activity of MG-63 and HOS cells suppressed by LINC00839 loss (Figure 4B). In addition, the promoting influence of si-LINC00839 on apoptosis in MG-63 and HOS cells was reversed after cotransfection with the miR-454-3p inhibitor (Figure 4C). Furthermore, interference of LINC00839 restricted the migration (Figure 4D) and invasion (Figure 4E) of MG-63 and HOS cells; however, cotransfection with the miR-454-3p inhibitor abolished these effects.
The c-Met overexpressing plasmid, pcDNA3.1-c-Met (pc-c-Met), or empty pcDNA3.1 plasmid was cotransfected with si-LINC00839 in MG-63 and HOS cells. Western blotting analysis revealed that transfection of pc-c-Met significantly increased c-Met protein expression in MG-63 and HOS cells (Figure 5A). Functional experiments indicated that elevated expression of c-Met neutralized the impacts of LINC00839 knockdown on proliferation (Figure 5B), apoptosis (Figure 5C), migration (Figure 5D), and invasion (Figure 5E) of MG-63 and HOS cells. Collectively, LINC00839 performed its pro-oncogenic roles in OS cells by regulating the activity of miR-454-3p/c-Met axis.
Inhibition of LINC00839 Suppresses OS Tumor Growth in vivo
Tumor xenograft assay was performed to test the biological role of LINC00839 on OS tumor growth in vivo. HOS cells stably transfected with sh-LINC00839 or sh-NC were subcutaneously injected into the flanks of nude mice. The tumor volume was apparently reduced in the mice injected with HOS cells stably expressing sh-LINC00839 (Figure 6A and B). The weight of the tumor xenografts was lower in the sh-LINC00839 group compared with that of the sh-NC group (Figure 6C). In addition, LINC00839 expression in the tumors from the sh-LINC00839 group was significantly decreased (Figure 6D). Furthermore, a significant increase of miR-454-3p (Figure 6E) and decrease of c-Met protein expression (Figure 6F) was observed in the tumors originating from sh-LINC00839 stably transfected HOS cells. Overall, the results of the tumor xenograft assay demonstrated that loss of LINC00839 attenuated OS tumor growth in vivo.
Discussion
In recent years, several studies have indicated that lncRNAs perform critical roles in regulating OS etiology and progression, and this has generated great interest in the scientific community.12,34,35 In view of their importance, studying the functions of lncRNAs in OS may offer new therapeutic targets and valuable prognostic biomarkers. The present study was done to examine the expression pattern of LINC00839 in OS and investigate its impact on the cellular processes of OS in vitro and in vivo. More importantly, the mechanisms responsible for its cancer-promoting activity in OS were explored in detail.
To our knowledge, this is the first study to explore the involvement of LINC00839 in the oncogenicity of OS. Initially, a total of 47 OS tissues and corresponding adjacent normal tissues were collected and used for qRT-PCR analysis to determine the expression of LINC00839. The data revealed high expression of LINC00839 in OS tissues and cell lines. The prognostic relevance of LINC00839 in OS was then evaluated using the Kaplan–Meier method. Patients with OS characterized a high LINC00839 expression manifested shorter overall survival relative to those patients with a low LINC00839 expression. Functionally, the inhibition of LINC00839 expression caused a significant reduction in OS cell proliferation, migration, and invasion in vitro. Furthermore, interference of LINC00839 expression attenuated OS tumor growth in vivo and induced apoptosis.
Additional work was performed to identify the molecular events involved in the pro-oncogenic activities of LINC00839, and therefore to acquire a comprehensive understanding of LINC00839 in OS. Because the function of lncRNAs depends on subcellular distribution,36 lncLocator, a lncRNA subcellular localization predictor, was utilized to forecast the cellular localization of LINC00839. LINC00839 was predicted to be primarily enriched in the cytoplasm, and this was further confirmed by subcellular fractionation experiment. It is well established that cytoplasmic lncRNA is likely to work as a ceRNA for miRNAs.21,23 LncRNAs possess miRNA responsive elements and can competitively bind to certain miRNAs and consequently decrease the repression of target mRNAs triggered by miRNAs, thus forming a complex lncRNA –miRNA-mRNA pathway.37 Accordingly, we suspect that LINC00839 works through such a ceRNA mechanism.
To verify our hypothesis, bioinformatics analyses were conducted to search for miRNAs that may bind to LINC00839 and miR-454-3p was found to be a potential candidate. Next, qRT-PCR analysis revealed that LINC00839 knockdown increased the expression of miR-454-3p in OS cells. Additionally, miR-454-3p was downregulated in OS, and there was an inverse relationship between the expression of miR-454-3p and LINC00839. Furthermore, luciferase reporter assay corroborated the direct binding of miR-454-3p to sequences of LINC00839. Moreover, a direct interaction between miR-454-3p and LINC00839 in OS cells was identified using RIP assay. Lastly, c-Met was demonstrated to be a direct target of miR-454-3p in OS cells which was under the control of LINC00839 through sponging of miR-454-3p. Altogether, we have identified a new LINC00839/miR-454-3p/c-Met network in OS cells.
MiR-454-3p is reported to be underexpressed in a variety of human cancers including OS.33 MiR-454-3p is regarded as a tumor-inhibiting miRNA in OS cells and is involved in the regulation of OS cell proliferation and invasion.33 C-Met, also known as MET, is a receptor for hepatocyte growth factor.38 It has an oncogenic role in OS progression by regulating a number of aggressive cell processes.39–42 In this study, our results indicated that miR-454-3p was able to directly target c-Met and negatively regulate its expression in OS cells. Furthermore, LINC0039 positively modulated c-Met expression in OS cells by acting as miR-454-3p sponge. Rescue experiments further demonstrated that the effect of LINC00839 deficiency on the biological processes of OS cells was abrogated, in part, by miR-454-3p inhibition or c-Met overexpression. Collectively, these results suggest that LINC00839 promotes the malignancy of OS cells by targeting the miR-454-3p/c-Met axis.
Conclusions
Our results identified the involvement of LINC00839 in OS progression. LINC00839 aggravated the OS cell phenotype by functioning as an miR-454-3p sponge and consequently increasing the expression of c-Met. Hence, the LINC00839/miR-454-3p/c-Met network may be a valuable target for OS treatment and prognosis.
Disclosure
The authors declare that they have no competing interests for this work.
References
1. Kager L, Tamamyan G, Bielack S. Novel insights and therapeutic interventions for pediatric osteosarcoma. Future Oncol. 2017;13(4):357–368. doi:10.2217/fon-2016-0261
2. Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8(10):705–718.
3. Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14(11):722–735.
4. Durfee RA, Mohammed M, Luu HH. Review of osteosarcoma and current management. Rheumatol Ther. 2016;3(2):221–243. doi:10.1007/s40744-016-0046-y
5. Lin YH, Jewell BE, Gingold J, et al. Osteosarcoma: molecular pathogenesis and iPSC modeling. Trends Mol Med. 2017;23(8):737–755. doi:10.1016/j.molmed.2017.06.004
6. Rasalkar DD, Chu WC, Lee V, Paunipagar BK, Cheng FW, Li CK. Pulmonary metastases in children with osteosarcoma: characteristics and impact on patient survival. Pediatr Radiol. 2011;41(2):227–236. doi:10.1007/s00247-010-1809-1
7. Yang J, Zhang W. New molecular insights into osteosarcoma targeted therapy. Curr Opin Oncol. 2013;25(4):398–406. doi:10.1097/CCO.0b013e3283622c1b
8. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
9. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi:10.1158/2159-8290.CD-11-0209
10. Greco S, Zaccagnini G, Perfetti A, et al. Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med. 2016;14(1):183. doi:10.1186/s12967-016-0926-5
11. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
12. Li Z, Dou P, Liu T, He S. Application of long noncoding RNAs in osteosarcoma: biomarkers and therapeutic targets. Cell Physiol Biochem. 2017;42(4):1407–1419. doi:10.1159/000479205
13. Jiang N, Wang X, Xie X, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi:10.1016/j.canlet.2017.06.009
14. Wang Z, Tan M, Chen G, Li Z, Lu X. LncRNA SOX2-OT is a novel prognostic biomarker for osteosarcoma patients and regulates osteosarcoma cells proliferation and motility through modulating SOX2. IUBMB Life. 2017;69(11):867–876. doi:10.1002/iub.1681
15. Deng R, Zhang J, Chen J. lncRNA SNHG1 negatively regulates miRNA1013p to enhance the expression of ROCK1 and promote cell proliferation, migration and invasion in osteosarcoma. Int J Mol Med. 2019;43(3):1157–1166. doi:10.3892/ijmm.2018.4039
16. Huang Q, Yang J, He X, Shi S, Xing S. LncRNA BDNF-AS is associated with the malignant status and regulates cell proliferation and apoptosis in osteosarcoma. Biosci Rep. 2018;38(6). doi:10.1042/BSR20181498
17. Zhou B, Li L, Li Y, Sun H, Zeng C. Long noncoding RNA SNHG12 mediates doxorubicin resistance of osteosarcoma via miR-320a/MCL1 axis. Biomed Pharmacother. 2018;106:850–857. doi:10.1016/j.biopha.2018.07.003
18. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi:10.1038/nrc1997
19. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
20. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
21. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334
22. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
23. Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 2019;20(22):5758.
24. He C, Chen H, Liu Y, et al. miR-106b-5p promotes cell proliferation and cell cycle progression by directly targeting CDKN1A in osteosarcoma. Exp Ther Med. 2020;19(5):3203–3210. doi:10.3892/etm.2020.8574
25. Chen J, Yan D, Wu W, Zhu J, Ye W, Shu Q. MicroRNA-130a promotes the metastasis and epithelial-mesenchymal transition of osteosarcoma by targeting PTEN. Oncol Rep. 2016;35(6):3285–3292. doi:10.3892/or.2016.4719
26. Li Z, Li Y, Wang N, Yang L, Zhao W, Zeng X. miR-130b targets NKD2 and regulates the Wnt signaling to promote proliferation and inhibit apoptosis in osteosarcoma cells. Biochem Biophys Res Commun. 2016;471(4):479–485. doi:10.1016/j.bbrc.2016.02.050
27. Wang W, Zhang L, Zheng K, Zhang X. miR-17-5p promotes the growth of osteosarcoma in a BRCC2-dependent mechanism. Oncol Rep. 2016;35(3):1473–1482. doi:10.3892/or.2016.4542
28. Liu R, Shen L, Qu N, Zhao X, Wang J, Geng J. MiR-19a promotes migration and invasion by targeting RHOB in osteosarcoma. Onco Targets Ther. 2019;12:7801–7808. doi:10.2147/OTT.S218047
29. Li X, Wang FS, Wu ZY, Lin JL, Lan WB, Lin JH. MicroRNA-19b targets Mfn1 to inhibit Mfn1-induced apoptosis in osteosarcoma cells. Neoplasma. 2014;61(3):265–273. doi:10.4149/neo_2014_034
30. Ni Z, Shang XF, Wang YF, Sun YJ, Fu DJ. Upregulated microRNA-301a in osteosarcoma promotes tumor progression by targeting CDC14A. Genet Mol Res. 2016;15(2):10–4238. doi:10.4238/gmr.15027807
31. Liang G, Duan C, He J, Ma W, Dai X. PTPN14, a target gene of miR-4295, restricts the growth and invasion of osteosarcoma cells through inactivation of YAP1 signalling. Clin Exp Pharmacol Physiol. 2020;47(7):1301–1310. doi:10.1111/1440-1681.13296
32. Zhang H, Zhang J, Meng F, et al. MicroRNA-93 promotes the tumorigenesis of osteosarcoma by targeting TIMP2. Biosci Rep. 2019;39(8).
33. Niu G, Li B, Sun J, Sun L. miR-454 is down-regulated in osteosarcomas and suppresses cell proliferation and invasion by directly targeting c-Met. Cell Prolif. 2015;48(3):348–355. doi:10.1111/cpr.12187
34. Wang JY, Yang Y, Ma Y, et al. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed Pharmacother. 2020;121:109627. doi:10.1016/j.biopha.2019.109627
35. Li Z, Yu X, Shen J. Long non-coding RNAs: emerging players in osteosarcoma. Tumour Biol. 2016;37(3):2811–2816. doi:10.1007/s13277-015-4749-4
36. Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol. 2017;1008:199–222.
37. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310. doi:10.3390/ijms19051310
38. Zhang J, Babic A. Regulation of the MET oncogene: molecular mechanisms. Carcinogenesis. 2016;37(4):345–355. doi:10.1093/carcin/bgw015
39. Naka T, Iwamoto Y, Shinohara N, Ushijima M, Chuman H, Tsuneyoshi M. Expression of c-met proto-oncogene product (c-MET) in benign and malignant bone tumors. Mod Pathol. 1997;10(8):832–838.
40. MacEwen EG, Kutzke J, Carew J, et al. c-Met tyrosine kinase receptor expression and function in human and canine osteosarcoma cells. Clin Exp Metastasis. 2003;20(5):421–430. doi:10.1023/A:1025404603315
41. Li X, Sun X, Wu J, Li Z. MicroRNA-613 suppresses proliferation, migration and invasion of osteosarcoma by targeting c-MET. Am J Cancer Res. 2016;6(12):2869–2879.
42. Wang K, Zhuang Y, Liu C, Li Y. Inhibition of c-Met activation sensitizes osteosarcoma cells to cisplatin via suppression of the PI3K-Akt signaling. Arch Biochem Biophys. 2012;526(1):38–43. doi:10.1016/j.abb.2012.07.003
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.