Back to Journals » Cancer Management and Research » Volume 11
Long noncoding RNA Linc00460 promotes breast cancer progression by regulating the miR-489-5p/FGF7/AKT axis
Authors Zhu Y, Yang L , Chong QY, Yan H, Zhang W, Qian W, Tan S, Wu Z, Lobie PE, Zhu T
Received 28 February 2019
Accepted for publication 27 April 2019
Published 1 July 2019 Volume 2019:11 Pages 5983—6001
DOI https://doi.org/10.2147/CMAR.S207084
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Yong Zhu,1 Leiyan Yang,1 Qing-Yun Chong,2 Hong Yan,3 Weijie Zhang,1 Wenchang Qian,1 Sheng Tan,1 Zhengsheng Wu,4 Peter E Lobie,5 Tao Zhu1
1Hefei National Laboratory for Physical Sciences at Microscale, the CAS Key Laboratory of Innate Immunity and Chronic Disease, School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, People’s Republic of China; 2Cancer Science Institute of Singapore and Department of Pharmacology, National University of Singapore, Singapore, Singapore; 3Department of Pathology, Anhui Provincial Cancer Hospital, The First Affiliated Hospital of University of Science and Technology of China, Hefei, Anhui, People’s Republic of China; 4Department of Pathology, Anhui Medical University, Hefei, Anhui 230032, People’s Republic of China; 5Tsinghua-Berkeley Shenzhen Institute, Tsinghua University, Shenzhen, Guangdong, People’s Republic of China
Purpose: Evidence indicates that long noncoding RNAs (lncRNA) possess important roles in various cellular processes and that dysregulation of lncRNAs promotes tumor progression. However, the expression patterns and biological functions of many specific lncRNAs in breast cancer remain to be determined.
Methods: Quantitative real-time polymerase chain reaction was performed to detect Linc00460, miR-489-5p and FGF7 expression. Protein levels were determined using Western blot. MTT and colony formation assay were used to measure cell proliferation. Transwell assays were conducted to determine cell migration and invasion. Luciferase reporter assays were carried out to assess the interaction between miR-489-5p and Linc00460 or FGF7. Biotin pull-down assay was used to detect the direct interaction between miR-489-5p and Linc00460. In vivo experiments were performed to measure tumor formation and lung metastasis.
Results: We demonstrated that lncRNA Linc00460 was upregulated in breast cancer, and its expression level was positively associated with lymphatic metastasis and poor overall survival. Forced expression of Linc00460 increased, whereas Linc00460 silencing decreased, breast cancer cell viability, migration and invasion both in vitro and in vivo. Linc00460 was identified as a direct target of miR-489-5p, which further targeted FGF7 and exerted oncogenic functions in breast cancer. Mechanistically, Linc00460 served as a competing endogenous RNA of FGF-7 mRNA by sponging miR-489-5p, resulting in upregulated FGF7 expression and AKT activity. Notably, forced expression of miR-489-5p abrogated Linc00460-mediated oncogenic behavior and activation of the FGF7-AKT pathway in breast cancer cells.
Conclusion: We have demonstrated that Linc00460 promotes breast cancer progression partly through the miR-489-5p/FGF7/AKT axis.
Keywords: Linc00460, miR-489-5p, breast cancer, FGF7, AKT
Introduction
Breast cancer is the most prevalent female malignancy.1 Despite significant progress in the diagnosis and treatment of breast cancer, the mortality of patients with advanced breast cancer remains high, being mainly attributed to frequent metastasis and recurrence.2,3
Long noncoding RNAs (lncRNAs) are defined as transcribed RNA molecules of more than 200 nucleotides with no protein-coding capacity and account for the majority of the transcribed genome.4 LncRNAs have been increasingly recognized as key players in cancer development and progression through regulation of cell proliferation, survival, metastasis, senescence, self-renewal, stem cell pluripotency and drug resistance.5,6,7,8,9,10 Recent studies have demonstrated that lncRNAs function by binding to specific target DNA, RNA and/or protein.11 Nevertheless, the clinical relevance and underlying mechanisms of lncRNAs have not been fully understood in breast cancer. Linc00460 (ID in NCBI database: 728192), a newly identified lncRNA, was reported to be dysregulated in various cancer types, including parapharyngeal schwannoma, renal cell carcinoma, head and neck squamous cell carcinoma and breast cancer.12,13,14 Repression of Linc00460 significantly inhibited the proliferation of esophageal and nasopharyngeal carcinoma cells.15,16 Furthermore, several cancer-related transcriptional factors, such as MYC, FOS, CTCF and JUN, have been suggested to regulate the expression level of Linc00460.17 Therefore, we sought to investigate the functional role of Linc00460 in breast cancer.
In the present study, we observed the increased expression of Linc00460 in breast cancer compared to the benign breast tissues. Moreover, Linc00460 expression in the breast cancer cohort was associated with lymph node metastasis and worse survival outcome. Linc00460 depletion decreased breast cancer cell proliferation, migration and invasion, and inhibited tumor growth and lung metastasis in vivo. Mechanistically, our study suggests that Linc00460 sponges endogenous miRNA-489-5p to activate FGF7-AKT signaling, thereby promoting breast cancer progression.
Materials and methods
Breast cancer specimens and cell lines
Forty-two breast cancer tissues and 15 benign mammary tissues were collected from patients who underwent surgery at the First Affiliated Hospital of Anhui Medical University (Hefei, Anhui, People’s Republic of China) between 2009 and 2010. Specimens were transported in liquid nitrogen and stored at –80°C until used. The inclusion criteria were as follows: patients had no history of prior therapy with anticancer drugs or radiotherapy; clinical information of each patient including age, tumor size, tumor stage and lymphatic metastasis was available and written informed consent was obtained from all patients. The study protocol regarding the use of clinical samples was reviewed and approved by the Biomedical Ethics Committee of Anhui Medical University and conducted in accordance with the Declaration of Helsinki. Data from GSE20711 and GSE6532 in the GEO database were extracted to analyze the expression patterns of Linc00460 and FGF7 in the breast cancer tissues. Breast cancer cell lines (MCF-7, BT-474, MDA-MB-231 and BT-549) and a mammary epithelial cell line (MCF-10A) used in this study were purchased from the American Type Culture Collection (Rockville, MD, USA). HMEC-hTERT cells were a gift of Prof. William Hahn, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts, United States of America. SUM159 and SUM149 cells were obtained from Asterand.18 Cell lines used in this study have been authenticated. MCF-7 and BT-474 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (Gibco) at 37°C with 5% CO2. BT-549 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum and 0.01 mg/mL human recombinant insulin (Sigma) at 37°C with 5% CO2. MDA-MB-231 cells were cultured in Leibovitz’s15 medium supplemented with 10% fetal bovine serum at 37°C in the absence of CO2. SUM159 and SUM149 cells were cultured in Ham F-12 (Invitrogen) supplemented with 5% fetal bovine serum, 5 µg/mL insulin and 1 µg/mL hydrocortisone (Sigma) at 37°C with 5% CO2. Cell culture medium for HMEC-TERT and MCF-10A cells was prepared as previously described.19
RNA extraction and qRT-PCR analysis
Total RNA was isolated from cells and clinical specimens using Trizol (Invitrogen, Carlsbad, CA) and converted to cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific Bio) following the manufacturer’s instructions. The resulting cDNA samples were analyzed by qPCR prepared using SYBR Premix Ex Taq Kit (Takara) and run on Stratagene Mx3000P (Agilent Technology). The expression levels of mRNAs and miRNAs were normalized to the expression level of GAPDH and U6 snRNA, respectively. The sequences of primers used are listed in Table S1.
Lentivirus production and stable cell line construction
The full-length Linc00460 DNA fragment was cloned and inserted into pSin vector (pSin) according to the manufacturer’s instructions. shRNAs targeting Linc00460 were synthesized and cloned into pLKO.1 vector. All primers and shRNA sequences are listed in Table S1. Lentivirus production and transduction were performed as previously described.20,21 Forty-eight hours after viral transduction, puromycin was used for selection of stably transduced cells.
MTT assays
Cells were seeded into 96-well plates at 1,000 cells per well 48 hrs after transfection or the indicated treatments. Cells were cultured for 5 days and cell viability was determined using the MTT reagent (Promega). Absorbance at 570 nm was measured after 2 hrs of incubation with the MTT reagent.
Colony formation assays
Cells were seeded into 6-well plates at 1,000 cells per well 48 hrs after transfection or the indicated treatments. After 10 days of culture, cells were washed with PBS, fixed with 10% formalin and stained with 0.5% crystal violet (Sigma). Representative images were used for the counting of respective colony numbers in each well.
Migration and invasion assays
Cells were seeded into the upper chamber of a 24-well transwell insert (8-μm pores, BD Biosciences, San Jose, CA) at 105 cells per well 48 hrs after transfection or the indicated treatments. Complete medium was added into the lower chamber. After 24–48 hrs, the remaining cells in the upper chamber were gently wiped off with a cotton swab. Cells migrated through the pores in the membrane to the lower surface of the chamber were fixed with 90% ethyl alcohol for half an hour and stained with 0.1% crystal violet. Images of the migrated cells were taken in three randomly chosen fields and the number of cells in each field was counted. The protocol of invasion assay is similar to migration except that the upper chambers were precoated with Matrigel (BD Biosciences) which was diluted by the FBS free medium.
Transfection and luciferase reporter assays
Full-length Linc00460 and 3’UTR of FGF7 were amplified and cloned separately into psiCHECK-2 vector. Linc00460 with mutated miR-489-5p-binding site (Linc00460-mut) was obtained by using specific primers as stated in Table S1. 5×104 MDA-MB-231 or MCF-7 cells were seeded into each well of 24-well plates 24 hrs before transfection. miR-489-5p mimics, miR-489-5p inhibitor or the respective scrambled oligonucleotides control was co-transfected with 0.1 µg of empty psiCHECK-2 vector control or psiCHECK-2 vectors containing Linc00460, Linc00460-mut or FGF7 3’UTR into the cells using Lipofectamine 2000 (Invitrogen). Cells were harvested and lysed 48 hrs after transfection, the luciferase activities were determined by Dual-Luciferase® Reporter Assay System (Promega).
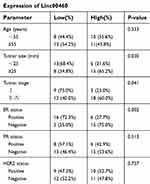 |
Table 1 Correlation of Linc00460 expression with clinicopathological features in 42 breast cancer patients |
Biotin pull-down assay
The detailed protocol was described previously.22 Briefly, sense or antisense biotin-labeled DNA oligomers targeting Linc00460 were incubated with lysates from MCF-7 cells. Streptavidin-coupled agarose beads (Invitrogen) were used to enrich the Linc00460 complex 1 hr after incubation. All steps were performed in RNase-free environment. Sequences of probes against Linc00460 are listed in Table S1.
Anti-AGO2 RNA immunoprecipitation
Anti-AGO2 RNA immunoprecipitation was performed as previously described.23 Briefly, breast cancer cells were lysed and the extract was incubated with anti-AGO2 antibody or anti-IgG (Santa Cruz, CA, USA). Protein G agarose beads (Invitrogen) was used to adhere to the immunoprecipitation product. Beads were washed and subjected to RNA extraction. Purified RNA was analyzed by qRT-PCR.
Protein extraction and Western blot
Total protein from cells was extracted by modified RIPA lysis buffer. Western blot was performed as previously described.24 Protein expression was analyzed by ImageQuant LAS4000 (GE Health). All antibodies used are listed in Table S2.
Xenograft and histological analyses
The protocol for animal experiments was approved by the Institutional Animal Care and Use Committee and followed the guideline of experimental animal welfare of the Laboratory Animal Ethical Committee at University of Science and Technology of China (USTCACUC1601015). Estradiol pellet (0.18 mg) was subcutaneously implanted into the dorsal skin of 4-week-old nude mice 1 week prior to injection of the MCF-7 cells. For the xenograft study, 5×106 MCF-7 control cells, or MCF-7 cells stably transfected with Linc00460, were mixed 1:1 with Matrigel (BD Biosciences) and injected into the second mammary fat pad of the nude mice (N=6). Tumor growth was determined by measuring the length (L) and width (W). Tumor volume was calculated using the formula Volume (mm3) = LW2 /2. After the last measurement, mice were sacrificed and tumors were collected, weighted, fixed and subjected to subsequent hematoxylin and eosin (H&E) staining analysis.
For tail vein injection, 2×106 MDA-MB-231 cells, which were stably transfected with Linc00460 shRNAs or empty vector, were injected into female BALB/c nude mice via the tail vein. Mice were sacrificed 2 months later and lungs were surgically resected for H&E staining as previously described.25
Statistical analysis
All the experiments were repeated at least three times. Student’s t-test or two-way ANOVA was used to analyze the difference between the two groups; chi-squared test was used to analyze the clinicopathological features of breast cancer patients and the frequency of lung metastases; Pearson correlation analysis was used to analyze the correlation between expressions of two genes; Kaplan–Meier’s analysis was used for prognostic analyses; univariate and multivariate Cox proportional hazards regression analyses were used to test for independent prognostic factors. All statistical analyses were performed using GraphPad Prism (San Diego, CA) and SPSS version 20.0 software and p<0.05 was considered as statistically significant.
Result
Elevated expression of Linc00460 in breast cancer predicts poor prognosis
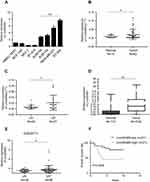
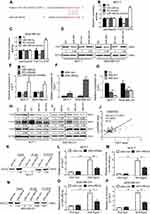
To evaluate the role of Linc00460 in breast cancer development and progression, we first assessed Linc00460 expression levels in two mammary epithelial cell lines (HMEC-HERT and MCF-10A) and 6 breast cancer cell lines (MCF-7, BT-474, SUM159, SUM149, MDA-MB-231 and BT-549) by qRT-PCR. The expression levels of Linc00460 were significantly higher in high invasive breast cancer cell lines (SUM159, SUM149, MDA-MB-231 and BT-549) as compared to the less invasive breast cancer cells (MCF-7, BT-474) and the normal mammary epithelial cell lines (Figure 1A). Next, we analyzed Linc00460 level in 42 breast cancer and 15 normal breast specimens. The expression levels of Linc00460 were found to be significantly elevated in breast cancer compared to the normal breast tissue (Figure 1B). As indicated in Table 1, further pathological examination demonstrated that higher expression of Linc00460 in breast cancer was correlated with larger tumor size (P=0.030) and advanced WHO stage (P=0.041). Linc00460 expression level was negatively correlated with ER positivity in breast cancer tissues (P=0.002). Notably, among the 42 breast cancer samples studied, 21 specimens obtained from patients with lymphatic metastasis (LN+) exhibited elevated levels of Linc00460 as compared to their lymphatic node metastasis-free (LN−) counterparts (Figure 1C). Consistently, increased expression of Linc00460 was observed in breast cancer compared to normal breast tissues in the TCGA BRCA cohorts with 1102 breast cancer tissues and 113 normal breast tissues (Figure 1D). A positive correlation between Linc00460 expression and lymphatic metastasis was also observed in the GSE20711 dataset (Figure 1E). Furthermore, patients with a higher expression level of Linc00460 exhibited a shorter overall survival rate. (Figure 1F). In addition, the prognostic value of Linc00460 was evaluated by univariate and multivariate Cox proportional hazards regression analyses. It was observed that Linc00460 was an independent prognostic factor for worse overall survival (HR =4.763, 95% CI: 1.135–19.985, P=0.033; Table 2). Collectively, these data suggest that Linc00460 is upregulated in breast cancer and is associated with a poor survival outcome.
 |
Table 2 Univariate and multivariate Cox proportional hazards regression analysis of Linc00460 for overall survival in breast cancer patients |
Linc00460 promotes the oncogenicity of breast cancer cells in vitro and in vivo
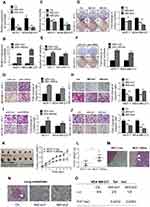
We next performed functional analyses of Linc00460 in breast cancer cells. MCF-7 (ER+) and MDA-MB-231 (ER-) carcinoma cells were stably transfected with Linc00460 shRNAs (460-sh1 or 460-sh2), Linc00460 expression plasmid, or their respective vector controls. The expression levels of Linc00460 in these cells were verified by qRT-PCR (Figure 2A and B). Depleted Linc00460 expression led to significantly reduced cell proliferation in both MCF-7 and MDA-MB-231 cells as determined by MTT and colony formation assays (Figure 2C and D). Conversely, forced expression of Linc00460 exerted opposite effects (Figure 2E and F). Furthermore, shRNA-mediated depletion of Linc00460 decreased, while the forced expression of Linc00460 increased, migration and invasion of MCF-7 and MDA-MB-231 cells in transwell assays (Figure 2G–J).
To determine the function of Linc00460 in breast cancer progression in vivo, 5×106 low Linc00460-expressing MCF-7 cells, stably transfected with either the Linc00460 expression plasmid or the empty vector, were injected orthotopically into the second mammary fat pad of female nude mice. Tumors derived from MCF-7 cells with forced expression of Linc00460 grew at a significantly faster rate with resultant mean tumor volume and weight approximately two-fold of those tumors derived from MCF-7-vector cells (Figure 2K and L). H&E staining of tumor sections revealed significant local invasion in tumors derived from MCF-7-Linc00460 cells, but not in tumors derived from MCF-7-vector cells (Figure 2M). To further assess the effect of Linc00460 on breast cancer metastasis, MDA-MB-231 cells transfected with either shRNA targeting Linc00460 or empty vector, were injected via the tail vein of 6-week-old nude mice. Depletion of Linc00460 expression produced a significant reduction in the number and size of lung micrometastases derived from metastatic MDA-MB-231 cells (Figure 2N). Furthermore, while all mice injected with MDA-MB-231-vector cells exhibited lung metastases (100%), a markedly lower frequency of lung metastases (only 25% and 12.5%) was observed in mice injected with MDA-MB-231-shLinc00460 cells (Figure 2O).
Linc00460 sponges miR-489-5p in breast cancer cells
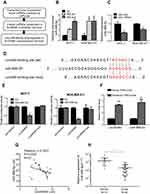
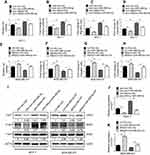
It has been reported that Linc00460 is abundantly expressed in the cytoplasm, suggestive of the possibility that Linc00460 acts as competitive endogenous RNA (ceRNA) or sponges of miRNAs in breast cancer.15,16 miRNAs may act as regulatory adaptors, through which lncRNAs, pseudogenes and messenger RNAs form regulatory crosstalk networks.26 Therefore, we hypothesized that Linc00460 may be a target of tumor suppressive miRNAs. We therefore searched the list of miRNAs predicted to target Linc00460 using miRcode (http://www.mircode.org/mircode/) and RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid). Eight of the identified predicted miRNAs have been reported to be tumor suppressors, of which three were significantly upregulated as a result of Linc00460 repression (data not shown) and only one of the three (miR-489-5p) was downregulated with forced expression of Linc00460 in breast cancer cells, suggestive that Linc00460 may be a target of miR-489-5p (Figure 3A–C). A potential miR-489-5p-binding site was also predicted on Linc00460 (Figure 3D). To verify the functionality of this binding site, Linc00460 with wild type or mutant miR-489-5p-binding sites were cloned into dual luciferase reporter vector (psiCHECK-2) and co-transfected with miR-489-5p or scrambled oligonucleotides into MCF-7 and MDA-MB-231 cells. Forced expression of miR-489-5p significantly decreased wild-type Linc00460-driven luciferase reporter activity but not that of the mutant Linc00460-driven luciferase reporter activity in both cell lines (Figure 3E). Furthermore, miR-489-5p, together with Linc00460, were enriched in the pull-down fraction of MCF-7 cells using biotin-conjugated probes against Linc00460 (Figure 3F). Concordantly, we observed a negative correlation between Linc00460 and miR-489-5p expression in breast cancer tissues by Pearson correlation analysis (Figure 3G). Additionally, expression levels of miR-489-5p were significantly lower in breast cancer compared to normal breast tissues (Figure 3H), further confirming that miR-489-5p functions as a tumor suppressor.
MiR-489-5p acts as a tumor suppressor in breast cancer
To verify the tumor suppressive role of miR-489-5p in breast cancer, MCF-7 and MDA-MB-231 cells were transfected with miR-489-5p mimics, single-strand miR-489-5p inhibitor or the respective scrambled oligonucleotides as the negative control (Figure 4A and B) and cell functions determined. MTT assays suggested that the forced expression of miR-489-5p decreased, while inhibition of miR-489-5p increased cell viability (Figure 4C and D). A similar result was observed in the colony formation assay (Figure 4E and F). Moreover, forced expression of miR-489-5p suppressed, while inhibition of miR-489-5p promoted the migration and invasion of MCF-7 and MDA-MB-231 cells in transwell assays (Figure 4G–J).
Linc00460 competes with FGF7 for binding of miR-489-5p
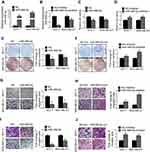
We further determined the functional targets of miR-489-5p in breast cancer. FGF7 was predicted to contain a conserved miR-489-5p-binding site using TargetScan (http://www.targetscan.org/) (Figure 5A).27 To verify that miR-489-5p targeted FGF7 mRNA, the 3’UTR of FGF7 was cloned into the psi-CHECK2 luciferase reporter plasmid and co-transfected with miR-489-5p mimics, miR-489-5p inhibitor or scrambled oligonucleotides into MCF-7 or MDA-MB-231 cells. Forced expression of miR-489-5p inhibited, while miR-489-5p antagonism increased FGF7 3’UTR-driven luciferase reporter activity in both MCF-7 and MDA-MB-231 cells (Figure 5B and 5C). We also examined whether miR-489-5p directly regulated FGF7 expression. Forced expression of miR-489-5p decreased, whereas miR-489-5p inhibition increased the mRNA and protein levels of FGF7 in MCF-7 and MDA-MB-231 cells (Figure 5D and E). As both Linc00460 and FGF7 are direct miR-489-5p targets, we next determined whether they competed for binding to miR-489-5p. Forced expression of Linc00460 increased FGF7 expression, whereas Linc00460 depletion decreased FGF7 expression in MCF-7 cells or MDA-MB-231 cells compared to control (Figure 5F–I). Elevated FGF7 expression has been reported to promote AKT signaling28, and we therefore examined AKT activity (phosphorylation) in these cells. We observed that the level of phosphorylated AKT was increased in MCF7 and MDA-MB-231 cells with forced expression of Linc00460 (Figure 5H and I). In contrast, phosphorylation of AKT was reduced, consistent with a decrease in FGF7 expression in MCF7 and MDA-MB-231 cells, upon Linc00460 depletion (Figure 5H and I). Additionally, analysis of the expression levels of Linc00460 and FGF7 from the GSE6532 dataset showed a significant positive correlation by Pearson correlation analysis (Figure 5J). We finally performed an anti-AGO2 RNA immunoprecipitation assay and observed Linc00460 overexpression reduced the enrichment of FGF7 mRNA on RISC by miR-489-5p in MCF7 and MDA-MB-231 cells (Figure 5K–P). Collectively, miR-489-5p inhibits FGF7-AKT signaling, in a manner that is inversely dependent on Linc00460 expression.
Linc00460 promotes oncogenicity through the miR-489-5p/FGF7/AKT axis
The FGF7-AKT axis has been reported to promote breast cancer progression.29,30,31 We therefore sought to determine whether the miR-489-5p/FGF-7/AKT axis mediated linc00460-mediated oncogenic behaviors in breast cancer cells. miR-489-5p mimics or scrambled oligonucleotides were transfected into MCF-7 cells to rescue Linc00460 overexpression reduced expression of miR-489-5p. As expected, forced expression of miR-489-5p abolished the growth, migration and invasion advantage conferred by Linc00460 overexpression in MCF-7 cells, as determined by MTT, colony formation and transwell assays (Figure 6A–D). Conversely, miR-489-5p inhibition rescued diminished growth, migration and invasion consequent to Linc00460 depletion in MDA-MB-231 cells (Figure 6E–H). Consistently, forced expression of miR-489-5p attenuated the Linc00460-mediated increase in FGF7 expression and AKT phosphorylation in MCF-7 cells (Figure 6I and J). In contrast, miR-489-5p inhibition rescued the Linc00460 depletion-mediated decrease in FGF7 expression and AKT phosphorylation in MDA-MB-231 cells (Figure 6I and K).
We further determined if FGF7 and its downstream signaling mediated the oncogenic functions of Linc00460. Linc00460 overexpressing breast cancer cell lines were transfected with FGF7 siRNAs or negative control. FGF7 depletion suppressed the Linc00460-mediated increase in p-AKT levels to levels similar to that of vector control cells (Figure 7A–C). Functionally, MTT and colony formation assays revealed that FGF7 silencing attenuated Linc00460-induced proliferation and/or survival of breast cancer cells to levels similar to that of vector control cells (Figure 7D–G). Furthermore, transwell assays showed that FGF7 depletion abrogated Linc00460-enhanced migratory and invasive capacities of breast cancer cells to levels similar to that of vector control cells (Figure 7H–K). Thus, FGF7 and its downstream AKT signaling mediate the oncogenic role of Linc00460.
Discussion
In this study, we investigated the expression pattern and clinical relevance of Linc00460 in breast cancer. We observed that Linc00460 was frequently upregulated in breast cancer. Forced expression of Linc00460 promoted viability, migration and invasion of breast cancer cells both in vitro and in vivo. Mechanistically, Linc00460 promoted breast cancer progression by sponging miR-489-5p and functioning as a competing endogenous RNA to FGF7 mRNA, consequently elevating FGF7 expression and enhancing its downstream AKT signaling.28
Recent studies have reported that the expression of Linc00460 is deregulated in renal cell carcinoma and head and neck squamous cell carcinoma.13 In addition, Linc00460 has also been reported to contribute to the development and progression of esophageal cancer and nasopharyngeal carcinoma.15,16 However, the underlying mechanism(s) by which Linc00460 promotes oncogenicity is not well delineated. We herein demonstrated that Linc00460 is a direct target of miR-489-5p. miR-489-5p has been reported to impair breast cancer proliferation and invasion by targeting the GSE1 coiled-coil protein.32 Interestingly, miR-489-3p, which originates from the same precursor, has been widely demonstrated to be a tumor suppressor, with functions in colorectal cancer, bladder cancer and osteosarcoma amongst other cancers.33–35 miR-489-3p has been reported to target histone deacetylase 7 (HDAC7) to suppress tumor growth and invasion in colon cancer.33 It has also been reported that the oncogenic KRAS/NF-kappaB/YY1 axis repressed miR-489-3p expression, leading to pancreatic cancer metastasis.36 In this study, we further investigated the functionality of miR-489-5p. In contrast to Linc00460, miR-489-5p was observed to be dramatically downregulated in breast cancer as compared to benign breast tissues. Functionally, we observed that miR-489-5p significantly inhibited the proliferation, migration and invasion of breast cancer cells, acting as a potent tumor suppressor.
Herein, FGF7 was identified as a target of miR-489-5p, competing with Linc00460 for miR-489-5p binding. FGF7 is a mesenchyme-specific heparin-binding growth factor that binds FGF receptor 2 (FGFR2) to regulate numerous cellular processes.37,38 Signaling pathways activated downstream of FGF7/FGFR2 include RAS-mitogen-activated protein kinase, Phospholipase C-γ, phosphoinositide3-kinase and Janus kinase/signal transducer and activator of transcription.28,39,40 Activation of FGF7/FGFR2 signaling has been reported to contribute to development and progression of gastric cancer, prostate cancer, lung adenocarcinoma and breast cancer.37,41–43 In the current study, we have demonstrated that increased Linc00460 expression inhibits miR-489-5p-mediated downregulation of FGF7 expression and is accompanied by consequent activation of FGF7-AKT pathway in breast cancer cells, revealing a novel cellular mechanism regulating the FGF7 pathway.
Linc00460 expression has previously been reported to be regulated by several well-known cancer-related transcription factors such as MYC, FOS, CTCF and JUN via bioinformatics prediction.17 Together with the prediction from rVista 2.0 (https://rvista.dcode.org/instr_rVISTA.html), we studied the effect of MYC, SP1, FOS transcription factors on the modulation of Linc00460 expression. However, we did not observe a significant change in the expression level of Linc00460 upon the depletion of these transcription factors. Therefore, the mechanism responsible for the upregulation of Linc00460 in breast cancer remains to be further investigated.
Conclusion
We demonstrated that Linc00460 was increased in expression in breast cancer and that the Linc00460/miR-489-5p/FGF7/AKT axis promoted proliferation, migration and invasion in breast cancer. A higher expression level of Linc00460 was prognostic for a shorter overall survival rate in breast cancer patients. Therefore, Linc00460 is a potential novel diagnostic biomarker for breast cancer prognosis.
Acknowledgments
This work was supported by The National Key Scientific Programme of China (2016YFC1302305), The National Natural Science Foundation of China (81672615, 81472494, 81672609), and Shenzhen Development and Reform Commission Subject Construction Project [2017] 1434.
Author contributions
All authors contributed toward data analysis, drafting and revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Disclosure
Profs. Peter E. Lobie and Tao Zhu consult for, and have equity interest in, Wuhan LongKe Ltd. Prof. Peter E. Lobie consults for AUM Biosciences Pte Ltd. and holds equity in Sinotar Pharmaceuticals Ltd., outside the submitted work. The authors report no other conflicts of interest in this work.
References
1. Bray F, McCarron P, Parkin DM. The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res. 2004;6(6):229–239.
2. Karamouzis MV, Papavassiliou AG. Targeting insulin-like growth factor in breast cancer therapeutics. Crit Rev Oncol Hematol. 2012;84(1):8–17.
3. Wood SL, Westbrook JA, Brown JE. Omic-profiling in breast cancer metastasis to bone: implications for mechanisms, biomarkers and treatment. Cancer Treat Rev. 2014;40(1):139–152.
4. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38.
5. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463.
6. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159.
7. Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–792.
8. Puvvula PK, Desetty RD, Pineau P, et al. Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nat Commun. 2014;5:5323.
9. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21.
10. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641.
11. Zhang Y, Tao Y, Liao Q. Long noncoding RNA: a crosslink in biological regulatory network. Brief Bioinform. 2017;19(5):930–945.
12. Carroll C, Jagatiya M. A rare case of a parapharyngeal schwannoma — an incidental finding. Int J Oral Maxillofac Surg. 2017;46:281.
13. Wang ZL, Li B, Piccolo SR, et al. Integrative analysis reveals clinical phenotypes and oncogenic potentials of long non-coding RNAs across 15 cancer types. Oncotarget. 2016;7(23):35044–35055.
14. Wang L, Li J, Zhao H, et al. Identifying the crosstalk of dysfunctional pathways mediated by lncRNAs in breast cancer subtypes. Mol Biosyst. 2016;12(3):711–720.
15. Liang Y, Wu Y, Chen X, et al. A novel long noncoding RNA linc00460 up-regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci Rep. 2017;37(5): BSR20171019.
16. Kong YG, Cui M, Chen SM, Xu Y, Xu Y, Tao ZZ. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84.
17. Cao W, Liu JN, Liu Z, et al. A three-lncRNA signature derived from the atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94–101.
18. Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527.
19. Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268.
20. Sankaran VG, Menne TF, Scepanovic D, et al. MicroRNA-15a and -16-1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. P Natl Acad Sci USA. 2011;108(4):1519–1524.
21. Sun LC, Song LB, Wan QF, et al. cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res. 2015;25(4):429–444.
22. Zhang P, Cao L, Fan P, Mei Y, Wu M. LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep. 2016;17(8):1204–1220.
23. Wang WX, Wilfred BR, Hu YL, Stromberg AJ, Nelson PT. Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. Rna. 2010;16(2):394–404.
24. Liu Y, Chen CY, Qian PX, et al. Gd-metallofullerenol nanomaterial as non-toxic breast cancer stem cell-specific inhibitor. Nat Commun. 2015;6: 5988.
25. Qian P, Banerjee A, Wu ZS, et al. Loss of SNAIL regulated miR-128-2 on chromosome 3p22.3 targets multiple stem cell factors to promote transformation of mammary epithelial cells. Cancer Res. 2012;72(22):6036–6050.
26. Su X, Xing J, Wang Z, Chen L, Cui M, Jiang B. microRNAs and ceRNAs: RNA networks in pathogenesis of cancer. Chin J Cancer Res. 2013;25(2):235–239.
27. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233.
28. Lei HP, Deng CX. Fibroblast growth factor receptor 2 signaling in breast cancer. Int J Biol Sci. 2017;13(9):1163–1171.
29. Lyakhovich A, Aksenov N, Pennanen P, et al. Vitamin D induced up-regulation of keratinocyte growth factor (FGF-7/KGF) in MCF-7 human breast cancer cells. Biochem Bioph Res Co. 2000;273(2):675–680.
30. Turczyk L, Kitowska K, Mieszkowska M, et al. FGFR2-driven signaling counteracts tamoxifen Effect on ERalpha-positive breast cancer cells. Neoplasia. 2017;19(10):791–804.
31. Zang XP, Siwak DR, Nguyen TX, Tari AM, Pento JT. KGF-induced motility of breast cancer cells is dependent on Grb2 and Erk1,2. Clin Exp Metastas. 2004;21(5):437–443.
32. Chai P, Tian J, Zhao D, et al. GSE1 negative regulation by miR-489-5p promotes breast cancer cell proliferation and invasion. Biochem Biophys Res Commun. 2016;471(1):123–128.
33. Gao S, Liu H, Hou S, et al. MiR-489 suppresses tumor growth and invasion by targeting HDAC7 in colorectal cancer. Clin Transl Oncol. 2018; 20(6):703-712.
34. Li J, Qu W, Jiang Y, et al. miR-489 suppresses proliferation and invasion of human bladder cancer cells. Oncol Res. 2016;24(6):391–398.
35. Liu Q, Yang G, Qian Y. Loss of MicroRNA-489-3p promotes osteosarcoma metastasis by activating PAX3-MET pathway. Mol Carcinog. 2017;56(4):1312–1321.
36. Yuan P, He XH, Rong YF, et al. KRAS/NF-kappaB/YY1/miR-489 signaling axis controls pancreatic cancer metastasis. Cancer Res. 2017;77(1):100–111.
37. Huang TT, Wang L, Liu D, et al. FGF7/FGFR2 signal promotes invasion and migration in human gastric cancer through upregulation of thrombospondin-1. Int J Oncol. 2017;50(5):1501–1512.
38. Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8(3):235–253.
39. Knights V, Cook SJ. De-regulated FGF receptors as therapeutic targets in cancer. Pharmacol Therapeut. 2010;125(1):105–117.
40. Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth F R. 2005;16(2):179–186.
41. Kwabi-Addo B, Ozen M, Ittmann M. The role of fibroblast growth factors and their receptors in prostate cancer. Endocr-Relat Cancer. 2004;11(4):709–724.
42. Yamayoshi T, Nagayasu T, Matsumoto K, Abo T, Hishikawa Y, Koji T. Expression of keratinocyte growth factor/fibroblast growth factor-7 and its receptor in human lung cancer: correlation with tumour proliferative activity and patient prognosis. J Pathol. 2004;204(1):110–118.
43. Penaultllorca F, Bertucci F, Adelaide J, et al. Expression of Fgf and Fgf receptor genes in human breast-cancer. Int J Cancer. 1995;61(2):170–176.
Supplementary materials
 |
Table S1 Oligomers used in this study |
 |
Table S2 Antibodies used in this study |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.