Back to Journals » OncoTargets and Therapy » Volume 13
Long Noncoding RNA HIF1A-AS2 Promotes Non-Small Cell Lung Cancer Progression by the miR-153-5p/S100A14 Axis
Authors Zhang W, Liu K, Pei Y, Tan J, Ma J, Zhao J
Received 16 May 2020
Accepted for publication 4 August 2020
Published 26 August 2020 Volume 2020:13 Pages 8715—8722
DOI https://doi.org/10.2147/OTT.S262293
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Weiqiang Zhang, Keqiang Liu, Yingxin Pei, Jian Tan, Jingbo Ma, Jing Zhao
Department of Thoracic Surgery, The 7th Medical Center of PLA General Hospital, Beijing 100700, People’s Republic of China
Correspondence: Jing Zhao
Department of Thoracic Surgery, The 7th Medical Center of PLA General Hospital, No. 5, Nanmencang Street, Dongcheng District, Beijing 100700, People’s Republic of China
Email [email protected]
Background: Long noncoding RNA (lncRNA) plays a critical role in initiating lung cancer. This study aims to research the function and mechanism of lncRNA HIF1A-AS2 in regulating non-small cell lung cancer (NSCLC) progression.
Methods: qRT-PCR was used to analyze gene expression. The CCK-8 assay was performed to detect cell proliferation. The Transwell assay was conducted to examine cell migration and invasion. A Caspase3 activity detection kit was utilized to analyze apoptosis. The luciferase reporter assay was carried out to research interactions of HIF1A-AS2, miR-153-5p and S100A14.
Results: HIF1A-AS2 expression was raised in NSCLC tissues and cell lines. The HIF1A-AS2 level was increased in advanced NSCLC tumor tissues. High HIF1A-AS2 expression was related to poor prognosis. HIF1A-AS2 knockdown decreased proliferation, migration and invasion while promoting apoptosis. HIF1A-AS2 was the sponge for miR-153-5p, and miR-153-5p targeted S100A14. HIF1A-AS2 promoted S100A14 expression through regulating miR-153-5p.
Conclusion: The HIF1A-AS2/miR-153-5p/S100A14 axis plays a crucial role in promoting NSCLC progression.
Keywords: NSCLC, HIF1A-AS2, miR-153-5p, S100A14
Introduction
Lung cancer is a common cancer and a leading cause of cancer-associated deaths worldwide.1 About two million people are diagnosed with lung cancer every year in the world.1 Non-small cell lung cancer (NSCLC) accounts for over 80% of all types of lung cancer.2 In spite of some advances having been made in lung cancer treatment, the 5-year overall survival rate is still lower than 20%.3 Tumor metastasis makes it more difficult to heal lung cancer. Thus, elucidation of the underlying mechanism of lung cancer carcinogenesis is urgently required.
Long noncoding RNA (lncRNA) is a subgroup of noncoding RNAs with a length of over 200 nucleotides and limited protein-coding ability.4 In the past decade, lncRNAs have attracted a lot of attention. More and more evidence has demonstrated that lncRNAs are highly expressed in many human tissues and are correlated with diseases such as cancer.5,6 Several biological processes, including proliferation, migration and chemo-resistance, are affected by lncRNAs in cancer.7 For example, lncRNA LL22NC03-N64E9.1 contributes to lung cancer growth and may be a potential prognostic biomarker.8 LncRNA MIR22HG works as a tumor suppressor and inhibits gastric cancer development via negatively regulating the NOTCH2 pathway.9 In addition, lncRNA LUCAT1 enhances ovarian cancer growth and metastasis via positively regulating the miR-612/HOXA12 axis.10 Importantly, several lncRNAs have been identified as potential biomarkers for cancer diagnosis or prognosis.11
HIF1A-AS2 expression is positively correlated with the development of osteosarcoma, bladder cancer, breast cancer and colorectal cancer.12–15 The roles of HIF1A-AS2 in NSCLC have not been discovered. Our current study has shown that HIF1A-AS2 expression was upregulated in NSCLC tissues and its downregulation attenuated proliferation, migration and invasion while promoting apoptosis. Mechanistically, HIF1A-AS2 was the sponge for miR-153-5p and facilitated S100A14 expression. Taken together, our findings suggest that the HIF1A-AS2/miR-153-5p/S100A14 axis plays a pivotal role in promoting NSCLC progression.
Patients and Methods
Patients
Forty-seven NSCLC tissues and neighboring normal tissues were collected from the 7th Medical Center of PLA General Hospital (Beijing, China) and stored in liquid nitrogen. This study was approved by the Ethics Committee of the 7th Medical Center of PLA General Hospital (20181216019). All patients signed the informed consent forms. This study was conducted in accordance with the Declaration of Helsinki. Human samples that had received no chemotherapy or radiotherapy prior to surgery were included. Associations between HIF1A-AS2 level and clinicopathological characteristics of NSCLC patients are presented in Table 1.
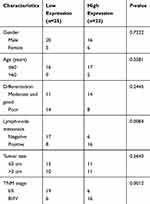 |
Table 1 Association Between HIF1A-AS2 Level and Clinicopathological Characteristics of NSCLC Patients |
Cell Culture and Transfection
NSCLC cell lines and the normal bronchial epithelial cell line BEAS-2B were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured with DMEM (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific). Small interfering RNAs (siRNAs) of HIF1A-AS2, miR-153-5p mimics, miR-153-5p inhibitors and negative controls were from Genepharma (Shanghai, China) and were transfected into cells using Lipofectamine 3000 (Thermo Fisher Scientific) following the manufacturer’s protocols. After 24 h of transfection, cells were used for the following experiments.
Quantitative RT-PCR
Total RNAs were extracted from NSCLC tissues using Trizol reagent (Invitrogen, Carlsbad, CA, USA). To detect the expression of miRNA-153-5p, reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific). PCR systems were prepared using the mirVana qRT-PCR miRNA Detection Kit (Thermo Fisher Scientific). For analysis of HIF1A-AS2 and S100A14 expression, RNAs were reverse transcribed into complementary DNA (cDNA) through a PrimeScript™ RT reagent Kit (Takara, Dalian, China), followed by qPCR detection using the SYBR Premix Ex Taq kit (Takara, Dalian, China). GAPDH or U6 acted as the normalized control. Relative expression was determined according to the 2−ΔΔCT method.
Cell Viability Assay
Proliferation or cell viability was detected using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Kumamoto, Japan) reagent according to the manufacturer’s protocols. The optical density at 450 nm was examined using an enzyme immunoassay analyzer (Thermo Fisher Scientific).
Transwell Migration and Invasion Assay
The Transwell assay was performed as described before.16 In brief, transfected cells were suspended in serum-free medium and seeded into the upper layer of the Transwell chamber (BD Biosciences; precoated with 50 μL Matrigel for the invasion assay). Then, 600 μL medium containing 20% FBS was added to the lower chamber. After 24 h, cells in the lower chamber were fixed in methanol, stained in 0.1% crystal violet and photographed under an inverted contrast microscope (CK2; Olympus, Japan).
Western Blot
Cells were lysed using radio-immunoprecipitation assay lysis buffer for 30 min and centrifuged at 13,000 g for 10 min. The protein concentration was subsequently measured using a bicinchoninic acid kit. Then, 20 μg protein was separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane. Subsequently, the membranes were blocked with 5% BSA and incubated with the diluted primary antibodies overnight. Then, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (1:10,000, ab7090; Abcam) for 1 h, followed by visualization using a SuperSignal West Pico Kit (Thermo Fisher Scientific).
RNA Immunoprecipitation (RIP)
The RIP assay was performed using the Magna RIP RNA-binding protein immunoprecipitation kit (MilliporeSigma) according to the manufacturer’s instructions. The cell lysates were incubated with magnetic beads and anti-Ago2 or IgG control. Then, the precipitated RNAs were isolated and subjected to qRT-PCR analysis.
Detection of Caspase3 Activity
Apoptosis was analyzed by detecting the activity of Caspase3 using a Caspase3 activity assay kit (Beyotime; Haimen, Jiangsu, China).
Dual-Luciferase Assay
miRDB and TargetScan tools were used for predictions among HIF1A-AS2, miR-153-5p and S100A14. For the luciferase reporter assay, the sequence of HIF1A-AS2 or S100A14 was cloned into pMIR-REPORT luciferase vector (Promega). Then, luciferase reporter vector and miR-153-5p mimics were transfected into NSCLC cells for 48 h. Finally, luciferase activity was measured by the Dual-Luciferase reporter assay system (Promega) according to the manufacturer’s instructions.
Statistical Analysis
All results were analyzed using GraphPad Prism7 and expressed as the mean ± SD. Differences were analyzed by Student’s t-tests for comparison of two groups or one-way analysis of variance for comparison of multiple groups. Correlations of expression in NSCLC tissues were detected by Pearson’s correlation coefficient. Overall survival rate was analyzed with the Kaplan–Meier method and the log rank test. P<0.05 was considered statistically significant.
Results
HIF1A-AS2 Presented High Levels in NSCLC Tissues
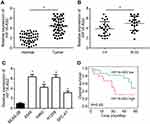
HIF1A-AS2 expression in NSCLC was explored. Through qRT-PCR analysis, it was found that HIF1A-AS2 expression was raised in NSCLC tissues compared to normal tissues (Figure 1A). Besides, HIF1A-AS2 expression was increased in NSCLC tissues with advanced stages (Figure 1B). Similarly, HIF1A-AS2 expression was upregulated in NSCLC cell lines compared to BEAS-2B cells (Figure 1C). Then, whether HIF1A-AS2 was a potential prognostic biomarker was assessed. Based on HIF1A-AS2 expression level (with the median value as the cut-off), NSCLC tissues were divided into two groups. Through Kaplan–Meier analysis, it was shown that high expression of HIF1A-AS2 was associated with a low survival rate (Figure 1D).
HIF1A-AS2 Knockdown Attenuated Growth, Survival and Metastasis
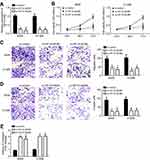
A549 and H1299 cells were used to perform the following experiments. Two siRNAs targeting HIF1A-AS2 were used. After transfection with siRNAs, HIF1A-AS2 expression was successfully decreased (Figure 2A). The CCK-8 assay showed that HIF1A-AS2 siRNA transfection induced a decrease in the proliferation rate (Figure 2B). Transwell assay results indicated that transfection of si-HIF1A-AS2 resulted in reduced migrated and invaded cells (Figure 2C and D). Besides, we found that HIF1A-AS2 knockdown increased the activity of Caspase3 (Figure 2E), suggesting that HIF1A-AS2 downregulation promotes apoptosis. Therefore, HIF1A-AS2 promotes NSCLC progression.
HIF1A-AS2 Was the Sponge for miR-153-5p
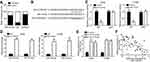
To explore the mechanism, we first examined the subcellular location of HIF1A-AS2 by qRT-PCR. The results showed that HIF1A-AS2 was mainly expressed in the cytoplasm of A549 cells (Figure 3A), implying that HIF1A-AS2 may be a miRNA sponge. We then identified the possible miRNA target through bioinformatics. We found miR-153-5p and constructed luciferase reporter vectors (Figure 3B). The luciferase reporter assay indicated that miR-153-5p mimics inhibited the activity of WT-HIF1A-AS2 vector (Figure 3C). Moreover, the RIP assay validated the interaction between HIF1A-AS2 and miR-153-5p (Figure 3D). HIF1A-AS2 knockdown promoted the expression of miR-153-5p in A549 and H1299 cells (Figure 3E). qRT-PCR analysis showed that miR-153-5p expression was negatively associated with HIF1A-AS2 level in NSCLC tissues (Figure 3F). These findings suggest that HIF1A-AS2 directly interacts with miR-153-5p in NSCLC.
miR-153-5p Targeted S100A14
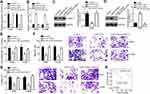
Subsequently, the downstream target of miR-153-5p was analyzed by bioinformatics. We identified S100A14 and constructed luciferase reporter vectors (Figure 4A). The luciferase reporter assay demonstrated the interaction between miR-153-5p and S100A14 (Figure 4B). In addition, miR-153-5p mimics repressed the mRNA levels of S100A14 in A549 and H1299 cells (Figure 4C and D). Notably, we observed that S100A14 expression was increased in NSCLC tissues compared to normal tissues according to TCGA database and qRT-PCR analysis (Figure 4E and F). Moreover, we found that the miR-153-5p level was negatively correlated with HIF1A-AS2 and S100A14 in NSCLC tissues (Figure 4G and H).
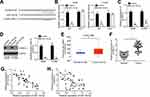 |
Figure 4 miR-153-5p targeted S100A14. (A) The predicted binding sites for S100A14 and miR-153-5p are presented. (B) Luciferase activity assay was performed to confirm the binding of S100A14 and miR-153-5p. (C and D) Expression level of S100A14 was analyzed by qRT-PCR and Western blot. (E) Expression level of S100A14 was analyzed using the TCGA database using the online tool UALCAN (http://ualcan.path.uab.edu/analysis.html). (F) Relative expression of S100A14 was measured in NSCLC tissues. (G and H) Expression correlations between HIF1A-AS2 and miR-153-5p or between miR-153-5p and S100A14 in 47 NSCLC tissues by qRT-PCR analysis. *P<0.05. |
HIF1A-AS2 Positively Affected NSCLC Progression Through the miR-153-5p/S100A14 Axis
We also observed that S100A14 expression was decreased after HIF1A-AS2 knockdown (Figure 5A) and this trend was reversed by miR-153-5p inhibitor transfection (Figure 5A). Besides, HIF1A-AS2 overexpression promoted S100A14 expression while miR-153-5p mimics abolished it (Figure 5B). Moreover, Western blot results further confirmed these findings (Figure 5C and D). Thus, HIF1A-AS2 inhibited miR-153-5p to upregulate S100A14 expression. Moreover, CCK-8 and Transwell assays showed that overexpression of S100A14 rescued the proliferation, migration and invasion of si-HIF1A-AS2 transfected NSCLC cells (Figure 5E–G), indicating that HIF1A-AS2 exerts roles through the miR-153-5p/S100A14 axis. Moreover, high expression of S100A14 was associated with a low survival rate (Figure 5H), indicating that S100A14 may be a potential prognostic marker.
Discussion
As a major problem for human health, NSCLC causes large numbers of deaths every year worldwide.1 Its underlying pathogenesis is unclear and effective therapeutic strategies are urgently lacking. In this study, we showed that HIF1A-AS2 expression was upregulated in NSCLC cells and was associated with clinical stage and prognosis. Loss-of-function assays demonstrated that HIF1A-AS2 knockdown suppressed proliferation and metastasis while promoting apoptosis in NSCLC. We also showed that HIF1A-AS2 was the sponge for miR-153-5p to enhance S100A14 expression. Our study disclosed that HIF1A-AS2 plays oncogenic roles in NSCLC.
Today, lncRNAs have been identified to regulate tumorigenesis, including in NSCLC.8 Dysregulation of lncRNA expression is usually observed in NSCLC.8 For example, lncRNA TP73-AS1 is overexpressed in NSCLC and promotes tumor development through targeting the miR-449a/EZH2 axis.17 LncRNA AFAP1-AS1 upregulation in NSCLC predicts an unsatisfactory prognosis and enhances cancer growth by inhibiting P21 levels.18 In addition, lncRNA SNHG12 silencing suppresses NSCLC proliferation and promotes cell death through sponging miR-138.19 HIF1A-AS2 has been reported to participate in several cancers, such as osteosarcoma, bladder cancer, breast cancer and colorectal cancer.12–15 Its function in NSCLC has yet to be described. Loss-of-function assays indicated that HIF1A-AS2 knockdown suppressed proliferation, migration and invasion while inducing apoptosis. Thus, HIF1A-AS2 is a critical oncogene in NSCLC.
The lncRNA–miRNA–mRNA regulatory axis has been widely acknowledged.17 There is evidence to support that lncRNAs are fine sponges for miRNAs in cancers.10,16 For example, lncRNA MT1JP interacts with miR-423-3p to upregulate Bim expression and regulates lung cancer progression.20 LncRNA DANCR sequesters miR-216a to promote lung cancer development.21 HIF1A-AS2 was also found to sponge several miRNAs, such as miR-129-5p, miR-548c-3p and miR-665.12,22,23 In our study, we demonstrated that HIF1A-AS2 interacted with miR-153-5p. We showed that miR-153-5p expression was upregulated after HIF1A-AS2 knockdown, and HIF1A-AS2 level was negatively correlated with miR-153-5p expression in NSCLC tissues. MiR-153-5p was reported to regulate acute myeloid leukemia and hepatocellular carcinoma.24,25 Our study, for the first time, identified the regulatory relationship between HIF1A-AS2 and miR-153-5p in NSCLC. Interestingly, we found that HIF1A-AS2 suppressed miR-153-5p expression. The mechanism through which HIF1A-AS2 affects miR-153-5p expression requires more investigation. Our findings suggested that miR-153-5p suppresses NSCLC progression. Subsequently, we analyzed the target of miR-153-5p through bioinformatics. We demonstrated that miR-153-5p directly targeted S100A14 and inhibited S100A14 expression in NSCLC. S100A14 has been previously shown to promote tumorigenesis.26,27 Its oncogenic roles in lung cancer have been demonstrated.26 Consistently, we also found that S100A14 was upregulated in NSCLC tissues. Moreover, we identified that S100A14 expression was regulated by the HIF1A-AS2/miR-153-5p axis. Finally, we proved that HIF1A-AS2 promoted NSCLC progression via facilitating S100A14 expression.
Conclusion
Our results show that HIF1A-AS2 works as a sponge for miR-153-5p to promote the expression of S100A14, contributing to NSCLC progression. However, there are some limitations in our research. For example, whether HIF1A-AS2 expression is associated with chemo-resistance or radio-resistance requires more investigation.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Hou J, Meng F, Chan LW, Cho WC, Wong SC. Circulating plasma microRNAs as diagnostic markers for NSCLC. Front Genet. 2016;7:193. doi:10.3389/fgene.2016.00193
3. Jones CM, Brunelli A, Callister ME, Franks KN. Multimodality treatment of advanced non-small cell lung cancer: where are we with the evidence? Curr Surg Rep. 2018;6(2):5. doi:10.1007/s40137-018-0202-0
4. Jathar S, Kumar V, Srivastava J, Tripathi V. Technological developments in lncRNA biology. Adv Exp Med Biol. 2017;1008:283–323.
5. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
6. Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol (Dordr). 2018;41(6):585–603. doi:10.1007/s13402-018-0406-4
7. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
8. Jing H, Qu X, Liu L, Xia H. A novel long noncoding RNA (lncRNA), LL22NC03-N64E9.1, promotes the proliferation of lung cancer cells and is a potential prognostic molecular biomarker for lung cancer. Med Sci Monit. 2018;24:4317–4323. doi:10.12659/MSM.908359
9. Li H, Wang Y. Long noncoding RNA (lncRNA) MIR22HG suppresses gastric cancer progression through attenuating NOTCH2 signaling. Med Sci Monit. 2019;25:656–665. doi:10.12659/MSM.912813
10. Yu H, Xu Y, Zhang D, Liu G. Long noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through regulation of miR-612/HOXA13 pathway. Biochem Biophys Res Commun. 2018;503(3):2095–2100. doi:10.1016/j.bbrc.2018.07.165
11. Zhao Z, Wang J, Wang S, Chang H, Zhang T, Qu J. LncRNA CCAT2 promotes tumorigenesis by over-expressed Pokemon in non-small cell lung cancer. Biomed Pharmacother. 2017;87:692–697. doi:10.1016/j.biopha.2016.12.122
12. Wang X, Peng L, Gong X, Zhang X, Sun R. LncRNA HIF1A-AS2 promotes osteosarcoma progression by acting as a sponge of miR-129-5p. Aging (Albany NY). 2019;11(24):11803–11813. doi:10.18632/aging.102448
13. Chen X, Liu M, Meng F, Sun B, Jin X, Jia C. The long noncoding RNA HIF1A-AS2 facilitates cisplatin resistance in bladder cancer. J Cell Biochem. 2019;120(1):243–252. doi:10.1002/jcb.27327
14. Wang Y, Zhang G, Han J. HIF1A-AS2 predicts poor prognosis and regulates cell migration and invasion in triple-negative breast cancer. J Cell Biochem. 2019;120(6):10513–10518. doi:10.1002/jcb.28337
15. Lin J, Shi Z, Yu Z, He Z. LncRNA HIF1A-AS2 positively affects the progression and EMT formation of colorectal cancer through regulating miR-129-5p and DNMT3A. Biomed Pharmacother. 2018;98:433–439. doi:10.1016/j.biopha.2017.12.058
16. Lu Q, Shan S, Li Y, Zhu D, Jin W, Ren T. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J. 2018;32(7):3957–3967. doi:10.1096/fj.201701237RR
17. Zhang L, Fang F, He X. Long noncoding RNA TP73-AS1 promotes non-small cell lung cancer progression by competitively sponging miR-449a/EZH2. Biomed Pharmacother. 2018;104:705–711. doi:10.1016/j.biopha.2018.05.089
18. Yin D, Lu X, Su J, et al. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer. 2018;17(1):92. doi:10.1186/s12943-018-0836-7
19. Wang X, Qi G, Zhang J, et al. Knockdown of long noncoding RNA small nucleolar RNA host gene 12 inhibits cell growth and induces apoptosis by upregulating miR-138 in nonsmall cell lung cancer. DNA Cell Biol. 2017;36(11):892–900. doi:10.1089/dna.2017.3830
20. Ma J, Yan H, Zhang J, Tan Y, Gu W. Long-chain non-coding RNA (lncRNA) MT1JP suppresses biological activities of lung cancer by regulating miRNA-423-3p/Bim Axis. Med Sci Monit. 2019;25:5114–5126. doi:10.12659/MSM.914387
21. Zhen Q, Gao LN, Wang RF, et al. LncRNA DANCR promotes lung cancer by sequestering miR-216a. Cancer Control. 2018;25(1):1073274818769849. doi:10.1177/1073274818769849
22. Guo X, Lee S, Cao P. The inhibitive effect of sh-HIF1A-AS2 on the proliferation, invasion, and pathological damage of breast cancer via targeting miR-548c-3p through regulating HIF-1alpha/VEGF pathway in vitro and vivo. Onco Targets Ther. 2019;12:825–834. doi:10.2147/OTT.S192377
23. Wu R, Ruan J, Sun Y, et al. Long non-coding RNA HIF1A-AS2 facilitates adipose-derived stem cells (ASCs) osteogenic differentiation through miR-665/IL6 axis via PI3K/Akt signaling pathway. Stem Cell Res Ther. 2018;9(1):348. doi:10.1186/s13287-018-1082-z
24. Shang J, Chen WM, Wang ZH, Wei TN, Chen ZZ, Wu WB. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp Hematol. 2019;70:42–54 e3. doi:10.1016/j.exphem.2018.10.011
25. Chen J, Huang X, Wang W, et al. LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR-153-5p/ARHGAP18 signaling axis. Aging (Albany NY). 2018;10(11):3371–3381. doi:10.18632/aging.101645
26. Ding F, Wang D, Li XK, et al. Overexpression of S100A14 contributes to malignant progression and predicts poor prognosis of lung adenocarcinoma. Thorac Cancer. 2018;9(7):827–835. doi:10.1111/1759-7714.12654
27. Zhu M, Wang H, Cui J, et al. Calcium-binding protein S100A14 induces differentiation and suppresses metastasis in gastric cancer. Cell Death Dis. 2017;8(7):e2938. doi:10.1038/cddis.2017.297
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.