Back to Journals » OncoTargets and Therapy » Volume 12
Long Noncoding RNA HCP5 Regulates Pancreatic Cancer Gemcitabine (GEM) Resistance By Sponging Hsa-miR-214-3p To Target HDGF
Authors Liu Y, Wang J, Dong L, Xia L, Zhu H, Li Z, Yu X
Received 10 July 2019
Accepted for publication 27 August 2019
Published 4 October 2019 Volume 2019:12 Pages 8207—8216
DOI https://doi.org/10.2147/OTT.S222703
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Yunfei Liu, Jiale Wang, Luo Dong, Li Xia, Hongwei Zhu, Zhiqiang Li, Xiao Yu
Department of Hepatobiliary and Pancreatic Surgery II, Third Xiangya Hospital, Central South University, Changsha 410006, China
Correspondence: Xiao Yu; Zhiqiang Li
Department of Hepatobiliary and Pancreatic Surgery II, Third Xiangya Hospital, Central South University, Changsha 410006, China
Tel +0731-88618576
Fax +0731-88618576
Email [email protected]; [email protected]
Background: Gemcitabine (GEM) is one of the most widely chemotherapy drugs in PC. However, the chemotherapy resistance always occurs after a period of treatment indicating poor prognosis. lncRNA may play an essential role in PC and serve as a prognosis biomarkers in PC with GEM-resistance. In our study, we aim to investigate the role of lncRNA HCP5 in PC.
Materials and methods: QRT-PCR detected the expression of lncRNA HCP5. The effects of knockdown lncRNA HCP5 on the proliferation, migration, invasion, cell apoptosis and autophagy were investigated in GEM-resistance PC cells. Bioinformatic analysis, luciferase reporter assay and RNA immunoprecipitation assay were performed to predict for potential miRNAs that can interact with lncRNA HCP5 and mRNAs that can interact with miR-214-3p.
Results: Our study revealed that lncRNA HCP5 expression was upregulated in PC tissues, especially increased expression in GEM-resistant PC tissues and GEM-resistant PC cells. Wound healing, Transwell assays, flow cytometry, Western blot, luciferase reporter assay and RNA immunoprecipitation (RIP) results demonstrated lncRNA HCP5 acted as a ceRNA to regulate GEM-resistance PC cells’ proliferation, invasion, migration, cell apoptosis and autophagy by targeting HDGF via miR-214-3p.
Conclusion: Our results revealed that lncRNA HCP5 is highly expressed in HCC, and development of GEM-resistance PC cells involving the processes of proliferation, invasive, migration, cell apoptosis and autophagy through the miR-214-3p/HDGF axis. Targeting lncRNA HCP5 may improve gemcitabine-based therapeutic efficacy.
Keywords: pancreatic cancer, lncRNA, gemcitabine, prognosis
Introduction
Pancreatic cancer (PC) is a highly malignant tumour of the digestive system. The incidence and mortality of PC are increasing year by year worldwide. 60% of patients with PC had distant metastasis at diagnosis, and only 10–15% of the patients had the chance to surgery.1–3 Currently, gemcitabine (GEM) is used as the first-line chemotherapy for advanced pancreatic cancer. However, most patients acquired drug resistance and the clinic efficacy of GEM is limited. Therefore, it is urgent to find new molecular therapeutic targets to reverse chemotherapy resistance and improve the prognosis of pancreatic cancer.
Long noncoding RNAs (lncRNAs) are a kind of noncoding RNA with more than 200 bp in length. Recent evidence indicated that lncRNAs function as oncogenes or tumour-suppressor genes are involved in various biological processes such as cell proliferation, cell metabolism, migration, invasion, cell cycle arrest, apoptosis and autophagy. Some studies also have confirmed that deregulation of lncRNAs were involved in tumour drug resistance, such as colorectal cancer, hepatocellular carcinoma, and breast cancer,4–8 and also showed lncRNAs were one of the most popular noncoding RNA that serve as prognosis biomarkers in GEM-resistance PC.
lncRNA human histocompatibility leukocyte antigen (HLA), HLA complex P5 (HCP5), is primarily found expressed in immune system cells and had a potential role in autoimmunity.9 Recent studies have shown that lncRNA HCP5 had been reported in some human cancers, such as thyroid carcinoma, triple-negative breast cancer, cervical cancer and colorectal cancer.10–13 Wenlong Wang et al utilized GSE16515, GSE15471 datasets and DAVID database to conduct functional enrichment analysis, and they demonstrated that MMP9/ITGB1-miR-29b-3p-lncRNA HCP5 competing endogenous RNA (ceRNA) subnetwork was linked to prognosis of pancreatic cancer.14 However, the roles of lncRNA HCP5 in PC have not been revealed. Therefore, we examined the expression of lncRNA HCP5 and investigated the biological function and mechanism in GEM-resistance PC cells.
In the present study, we found that lncRNA HCP5 expression was upregulated in PC tumour tissues, especially in GEM-resistant PC tissues and GEM-resistant PC cells. Cytological studies demonstrated that knockdown lncRNA HCP5 could affect PC cells’ proliferation, invasion, cell apoptosis and autophagy. Luciferase assay and RIP suggested that lncRNA HCP5 acted as a (competing endogenous RNA) ceRNA to regulate hepatoma-derived growth factor (HDGF) by sponging miR-214-3p. Our research revealed that lncRNA HCP5 may represent a new therapy target for GEM-resistant PC.
Materials And Methods
Clinical Specimens
A total of 28 PC tissue samples and matched normal tissue were collected From Third Xiangya Hospital, Central South University, January 2017 to September 2018. Gemcitabine (GEM) resistance definition: GEM treatment was not effective in PC patients or less than 6 months of previous gemcitabine treatment had progression. All tissue samples were cleaned and immediately stored in liquid nitrogen and stored until use. The study was approved by the Ethics Committee Third Xiangya Hospital of Central South University. All participants were informed and signed the informed consent form for this study.
Cell Lines And Culture Conditions
PANC-1 and SW 1990 were purchased from the Cell Collection Committee of the Chinese Academy of Sciences (Shanghai, China). All PC cells were incubated at the Cell Collection Committee recommended conditions. The gmcitabine-resistant cell lines PANC-1-GR and SW 1990-GR were established according to a previous study.15 miR-214-3p inhibitor and lentivirus containing shRNA-mediated lncRNA HCP5 downregulation and HDGF downregulation as well as their negative controls were purchased from RiboBio (Guangzhou, China). PANC-1-GR and SW 1990-GR were cultured in 6-well plates for 24 hrs and then were transfected with the indicated lentivirus shRNA-lncRNA HCP5 or miR-214-3p inhibitor using Lipofectamine 3000 (Invitrogen, California, USA) according to the manufacture’s protocol. The use of cell lines was approved by the ethics committee of the institutional review board (IRB) of The Third Xiangya Hospital, Central South University (IRB2018-S138).
RNA Extraction And Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen, CA, USA) from the Human PC tissues or cells. Total RNA was transcribed into cDNA by PrimeScript RT reagent Kit (Takara, Dalian, China) following with the manufacturer’s protocol. The expression of lncRNA HCP5, miR-214-3p and HDGF was measured by Fast-Start Universal SYBR Green Master (Rox) (Roche, Switzerland) following with manufacturer’s protocol. After that, real-time PCR was performed at the conditions: 95.0°C for 5 mins and 40 circles of 95.0°C for 30 s and 70°C for 60 s in a CFX96 Tm Real-Time System (Bio-Rad, California, USA). lncRNA HCP5, miR-214-3p, HDGF and endogenous reference primers were synthesized by Sangon Biotech (Shanghai, China). Each detection was replicated three times. Fold changes in expression of each gene were calculated by a comparative Ct method (2−∆∆Ct or −∆Ct).
CCK-8 Assay
Cell Counting Kit-8 (Sigma-Aldrich, St. Louis, MO, USA) was used to analyze cell viability. After transfection, PANC-1-GR and SW 1990-GR cells were seeded into 96-well plates cultured for 24 hrs and followed by incubation for 4 hrs. Finally, the cell proliferation assay was performed according to the manufacturer’s instructions.
Scratch And Transwell Assays
In vitro invasion and migration of PANC-1-GR and SW 1990-GR cells were measured by wound healing and Transwell assays. About 1×105 transfected cells were seeded into the upper chambers of each transwell. Medium containing 20% FBS was added to the bottom chamber and incubated at 37°C for 24 hrs, then the cells on the lower surface of the filter were fixed and stained, and migrated cells were measured by averaging the total numbers of cells from triplicate determinations.
Apoptosis Assay
Cell apoptosis assay was performed using flow cytometry after staining with Annexin V-labelled with FITC and PI detection kit (Life technologies, New York, NY, USA) based on manufacturer’s protocol, and cell apoptosis was analyzed by flow cytometer (BD Biosciences, New Jersey, USA).
Dual-Luciferase Reporter Assay
PANC-1-GR and SW 1990-GR cells were seeded into 24-well plates and co-transfected with the shRNA lncRNA HCP5 or miR-214-3p mimic plasmid or control vector. lncRNA HCP5 wild-type (WT), HDGF WT and mutant lncRNA HCP5 MUT 3′-UTR, HDGF MUT 3′-UTR were constructed. The plasmid (Promega, Fitchburg, WI, USA) expressing Renilla luciferase was co-transfected to control transfection efficiency after 48 hr transfection, luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. We used a dual-luciferase reporter assay system (Promega, Madison, WI, USA) and measured the luciferase activity and the normalized Renilla activity.
RNA-Binding Protein Immunoprecipitation (RIP)
PANC-1-GR and SW 1990-GR cells were co-transfected with shRNA lncRNA HCP5, miR-214-3p mimic plasmid or control vector, then total RNA was purified using RNeasy Mini Kit (QIAGEN) for further analysis, and detected by qRT-PCR.
In Vivo Metastasis Assay
We selected 4- to 8-week old BALB/c nude mice for in vivo tumour xenograft experiments. 100 mL of shRNA lncRNA HCP5 co-transfected into PANC-1-GR and SW 1990-GR cells (1×106) were subcutaneously injected into the upper back of BALB/c nude mice. The tumour size was measured weekly, as follows: W2×L/2. One month after the injection, the mice were euthanized to measure the tumour weight. Instructive notions with respect to caring for laboratory animals (which is released by the Ministry of Science and Technology of the People’s Republic of China in September 30, 2006.) were followed for the welfare of the animals. The laboratory animals were approved by the medical laboratory animal ethics committee of Third Xiangya Hospital, Central South University (AEC2018-S156).
Western Blot Analysis
The total tumour cells were homogenized and the total protein was extracted by RIPA buffer. And then the extraction and detection of the proteins were performed by Western blotting. Anti-HDGF antibody (1:1000), anti-LC3 I, LC3 II antibody (1:1000), anti-Beclin-1 antibody (1:1000), anti-P62 antibody (1:1000), anti-HDGF antibody (1:1000), anti-GAPDH antibody (1:2000) (Cell Signaling Technology, Boston, USA), then membranes were incubated with a secondary antibody (1:5000, Cell Signaling Technology, USA), and all protein samples were measured using a chemiluminescence (Bio-Rad, USA) imaging system.
Statistic Analysis
Continuous variables were expressed as means±SD. One-way ANOVA was performed for multiple comparisons using GraphPad Prism software, version 5.0 (GraphPad, La Jolla, CA, USA). Between differences two groups data were analyzed using the Student’s t-test. Pearson correlation analysis was conducted to determine the relationship between lncRNA HCP5, miR-214-3p and HDGF levels in PC specimens, and Kaplan–Meier curve and log-rank test were used for survival analysis. Statistical significance was considered at P<0.05.
Results
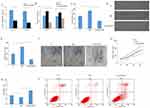
lncRNA HCP5 Is Upregulated In Patients And Cell Lines With PC Gemcitabine (GEM) Resistance
QRT-PCR results showed that the expression of lncRNA HCP5 in PC tissues was significantly upregulated compared with the normal tissues (P<0.001, Figure 1A). Moreover, compared with GEM-sensitive PC tissues, lncRNA HCP5 expression was significantly increased in GEM-resistant PC tissues (P<0.001, Figure 1B). Further, Kaplan–Meier curve and log-rank test in 28 PC patients evaluated that PC patients with high expression of lncRNA HCP5 indicated poorer survival rate (P<0.001, Figure 1C). We also detected the expression of lncRNA HCP5 in GEM-resistance PC cell lines (PANC-1-GR and SW 1990-GR)and their parental cell lines (PANC-1 and SW 1990), the expression of lncRNA HCP5 was significantly over-expression in PANC-1-GR and SW 1990-GR cells (P<0.05, Figure 1D).
Low-Expression lncRNA HCP5 Regulates PC Cells’ Proliferation, Invasive, Migration, Cell Apoptosis And Autophagy
To investigate the function of lncRNA HCP5 in PC, we performed lncRNA HCP5 knockdown experiments using specific shRNA into PANC-1-GR and SW 1990-GR. And then RT-PCR assay showed sh-lncRNA HCP5 efficiently suppressed the endogenous expression of lncRNA HCP5 in PANC-1-GR and SW 1990-GR cells (Figure 2A).
In addition, CCK8 assay, wound healing and transwell assays demonstrated that lncRNA HCP5 knockdown could significantly inhibit invasion and migration (Figure 2B–F). Furthermore, in vivo tumour xenografts were found that the average weight in the lncRNA HCP5 knockdown group was significantly smaller than the control group (Figure 2G). Flow cytometry and Western blot also showed depletion of lncRNA HCP5 remarkably increased cell apoptosis and regulated the cell autophagy of GEM-resistance PC cells (Figures 2H and I and 4E). These results indicate that depletion of lncRNA HCP5 promotes PC cell sensitization to gemcitabine (GEM) treatment.
lncRNA HCP5 Serves As A Sponge For miR-214-3p To Target HDGF In PC Cells
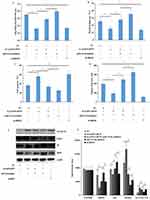
In general, mounting evidence had indicated the close link between lncRNA and miRNAs in the regulation of biological processes. To explore the mechanism of lncRNA HCP5 affects on PC growth and chemosensitivity. We searched online database DIANA-LncBase v2 (http://www.microrna.gr/LncBase) and starBase V3.0 (http://starbase.sysu.edu.cn/) to predict miRNAs interact with lncRNA HCP5. And found that 21 miRNAs interact with lncRNA HCP5. We performed a luciferase screening with 21 miRNA mimic to identify the miRNAs that bind sites of lncRNA HCP5. The luciferase activity of 6 miRNAs (miR-29b-3p, miR-214-3p, miR-140-5p, miR-3619-5p, miR-17-5p and miR-128-3p) was significantly decreased compared with the control group. In addition, we further confirmed that there were no significant alterations on luciferase activity in the cells cotransfected with mutant form MUT-lncRNA HCP5 (Figure 3A–C), and RIP assay results demonstrated that miR-214-3p was copurified with lncRNA HCP5 (Figure 3D). The expression levels of miR-214-3p in PANC-1-GR and SW 1990-GR cells transfected with sh-lncRNA HCP5 were upregulated compared with parental control (Figure 3E).
We used online databases (microT, miRmap, PITA, starbase, picTar, RNA22 and miRanda) to find potential target genes for miR-214-3p and take intersections by analyzing their results. We found that the binding sites of HDGF and miR-214-3p. Luciferase reporter gene assay showed that miR-214-3p directly bound to 3ʹ UTR of HDGF (Figure 3F). And then compared with the control group, the luciferase activity in co-transfected PANC-1-GR cells with HDGF and miR-214-3p-WT was significantly decreased. However, transfected HDGF and miR-214-3p-MUT did not change significantly (Figure 3G). These results confirmed that miR-214-3p directly target HDGF. Then, we detected the expression of HDGF mRNA in PANC-1-GR cells transfected with sh-lncRNA HCP5 or sh-lncRNA HCP5+ miR-214-3p inhibitor or miR-214-3p inhibitor or negative control by RT-PCR. The results showed that the expression of HDGF was significantly reduced in PANC-1-GR cells transfected with sh-lncRNA HCP5 and upregulated by sh-lncRNA HCP5+miR-214-3p inhibitor, miR-214-3p inhibitor (Figure 3H).
In addition, the expressions of miR-214-3p and HDGF were detected in 28 pairs of PC tumour tissues and adjacent normal tissues. The expressions of miR-214-3p were decreased in PC tumour tissues. Notably, miR-214-3p was significantly reduced in GEM-resistant PC tumour tissues (Figure 3I). HDGF expressions were increased in PC tumour tissues, and HDGF was significantly increased in GEM-resistant PC tumour tissues (Figure 3J). Correlation analysis showed that lncRNA HCP5 was negatively correlated with miR-214-3p, miR-214-3p was negatively correlated with HDGF and lncRNA HCP5 was positively correlated with HDGF (Figure 3K). In summary, these results demonstrated that lncRNA HCP5 serves as a sponge for miR-214-3p to target HDGF in PC cells.
LncRNA HCP5 acted as a ceRNA to regulate GEM-resistance PC cells’ proliferation, invasion, migration, cell apoptosis and autophagy by targeting HDGF via miR-214-3p
We investigated whether lncRNA HCP5 exerted its oncogenic role through regulating the expression of HDGF through direct binding miR-214-3p. miR-214-3p inhibitor could reverse sh-lncRNA HCP5 effect in PANC-1-GR cells’ proliferation, invasion, migration, cell apoptosis and autophagy (Figure 4A–D). Western blot showed that the protein levels of HDGF was similar in PANC-1-GR cells (Figure 4E). Furthermore, sh-lncRNA HCP5 could significantly decrease p62 and increase Beclin1, LC3 I/LC3 II, while miR-214-3p inhibitor significantly increased p62 and decreased Beclin1, LC3 I/LC3 II. However, miR-214-3p inhibitor significantly abolished decreased p62 and increased Beclin1, LC3 I/LC3 II induced by the knockdown of lncRNA HCP5 (Figure 4E and F). This indicated that lncRNA HCP5 low-expression vector can neutralize the effect of miR-214-3p inhibitor in GEM-resistance PC. In conclusion, these results demonstrated that lncRNA HCP5 contributed to GEM resistance by acting as a ceRNA to negatively modulate miR-214-3p expression and target HDGF to regulate PC cells’ proliferation, invasion, migration, cell apoptosis and autophagy.
Discussion
More and more studies have shown that lncRNA plays an important role in the development of tumorigenesis.16 Therefore, we chose lncRNA HCP5 for research. Some previous studies have examined that the expression of lncRNA was involved in the chemotherapy resistance initiation and progression of PC, for example, lncRNA GAS5 reverses EMT and tumour stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer.17 Long noncoding RNA LINC00346 promotes pancreatic cancer growth and gemcitabine resistance by sponging miR-188-3p to derepress BRD4 expression.18 lncRNA TUG1 promoted viability and is associated with gemcitabine resistance in pancreatic ductal adenocarcinoma.19 LncRNAAWPPH promotes osteosarcoma progression via activation of Wnt/beta-catenin pathway through modulating miR-93-3p/FZD7 axis. Long noncoding RNA CASC9 enhances breast cancer progression by promoting metastasis through the meditation of miR-215/TWIST2 signalling associated with TGF-beta expression.20,21 In our study, lncRNA HCP5 was identified to exert significant upregulation in GEM-resistance PC tissues and GEM-resistant PC cells. Similarly, lncRNA HCP5 overexpression displayed poorer overall survival compared with the PC patients with lower expression.
It is widely accepted that lncRNAs could function as a ceRNA binding to miRNAs and miRNAs can target mRNAs. We used online databases to find the potential target genes for lncRNA HCP5 and miR-214-3p. Luciferase reporter and RIP assay evidenced that miR-214-3p could interact with lncRNA HCP5. And HDGF could interact with miR-214-3p. RT-PCR results showed that the expression of ZEB1 mRNA was significantly upregulated in GEM-resistance PC cell and cancer tumour tissues. Correlation analysis showed that lncRNA HCP5 was negatively correlated with miR-214-3p, miR-214-3p was negatively correlated with HDGF and lncRNA HCP5 was positively correlated with HDGF. And miR-214-3p had been reported to regulate proliferation, migration and invasion in endometrial carcinoma cells, glioma cells, gastric cancer and osteosarcoma by directly targeting certain genes NEAT1, EZH2, RUNX3 and CADM1.22–26
Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion, and autophagy induction in pancreatic ductal adenocarcinoma (PDA) occurs as part of a broader transcriptional program that coordinates the activation of lysosome biogenesis and function, and nutrient scavenging, mediated by the MiT/TFE family of transcription factors.27,28 Subsequently, shRNA technology was used for the function of lncRNA HCP5 studies. CCK-8 assay showed that the proliferation of GEM-resistance PC cells after lncRNA HCP5 knockout was significantly inhibited. Flow cytometry revealed that the apoptosis rate of GEM-resistance PC cells increased after silencing. Wound healing and Transwell assays also demonstrated lncRNA HCP5 knockdown in PC GEM-resistance cells could significantly inhibit cells’ invasive, migration. As expected, our study indicated that lncRNA HCP5 plays a large role in pancreatic cancer cell proliferation inhibition and apoptosis induction by regulating gemcitabine sensitivity. Above all, these results confirmed that lncRNA HCP5 regulates GEM-resistance PC cells by targeting miR-214-3p/HDGF axis.
Conclusion
In conclusion, this study showed that lncRNA HCP5 contributed to GEM resistance and may act as a ceRNA to regulate HDGF gene expression by sponging miR-214-3p in PC. Cytological studies demonstrated that knockdown lncRNA HCP5 could affect PC cells’ proliferation, invasion, cell apoptosis and autophagy. Our results suggest that lncRNA HCP5 plays oncogene effects on PC and regulates GEM-resistance PC cells by targeting miR-214-3p/HDGF axis.lncRNA HCP5 may represent a new therapy target for GEM-resistant in PC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–1953. doi:10.1002/ijc.31937
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi:10.1016/S0140-6736(16)00141-0
4. Zhi Y, Abudoureyimu M, Zhou H, et al. FOXM1-mediated LINC-ROR regulates the proliferation and sensitivity to sorafenib in hepatocellular carcinoma. Mol Ther Nucleic Acids. 2019;16:576–588. doi:10.1016/j.omtn.2019.04.008
5. Li W, Dong X, He C, et al. LncRNA SNHG1 contributes to sorafenib resistance by activating the Akt pathway and is positively regulated by miR-21 in hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2019;38(1):183. doi:10.1186/s13046-019-1177-0
6. Wu J, Chen H, Ye M, et al. Downregulation of long noncoding RNA HCP5 contributes to cisplatin resistance in human triple-negative breast cancer via regulation of PTEN expression. Biomed Pharmacother. 2019;115:108869. doi:10.1016/j.biopha.2019.108869
7. Tong J, Ma X, Yu H, Yang J. SNHG15: a promising cancer-related long noncoding RNA. Cancer Manag Res. 2019;11:5961–5969. doi:10.2147/CMAR.S208054
8. Lou S, Xu J, Wang B, et al. Downregulation of lncRNA AFAP1-AS1 by oridonin inhibits the epithelial-to-mesenchymal transition and proliferation of pancreatic cancer cells. Acta Biochim Biophys Sin (Shanghai). 2019;51(8):814–825. doi:10.1093/abbs/gmz071
9. Liu Y, Helms C, Liao W, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4(3):e1000041. doi:10.1371/journal.pgen.1000041
10. Chen J, Zhao D, Meng Q. Knockdown of HCP5 exerts tumor-suppressive functions by up-regulating tumor suppressor miR-128-3p in anaplastic thyroid cancer. Biomed Pharmacother. 2019;116:108966. doi:10.1016/j.biopha.2019.108966
11. Wang L, Luan T, Zhou S, et al. LncRNA HCP5 promotes triple negative breast cancer progression as a ceRNA to regulate BIRC3 by sponging miR-219a-5p. Cancer Med. 2019,8(9):4389–4403. doi:10.1002/cam4.2335
12. Yang C, Sun J, Liu W, et al. Long noncoding RNA HCP5 contributes to epithelial-mesenchymal transition in colorectal cancer through ZEB1 activation and interacting with miR-139-5p. Am J Transl Res. 2019;11(2):953–963.
13. Yu Y, Shen HM, Fang DM, Meng Q-J, Xin Y-H. LncRNA HCP5 promotes the development of cervical cancer by regulating MACC1 via suppression of microRNA-15a. Eur Rev Med Pharmacol Sci. 2018;22(15):4812–4819. doi:10.26355/eurrev_201808_15616
14. Wang W, Lou W, Ding B, et al. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging (Albany NY). 2019;11(9):2610–2627. doi:10.18632/aging.101933
15. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
16. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.010
17. Liu B, Wu S, Ma J, et al. lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Mol Ther Nucleic Acids. 2018;13:472–482. doi:10.1016/j.omtn.2018.09.026
18. Shi W, Zhang C, Ning Z, et al. Long non-coding RNA LINC00346 promotes pancreatic cancer growth and gemcitabine resistance by sponging miR-188-3p to derepress BRD4 expression. J Exp Clin Cancer Res. 2019;38(1):60. doi:10.1186/s13046-019-1055-9
19. Yang F, Li X, Zhang L, Cheng L, Li X. LncRNA TUG1 promoted viability and associated with gemcitabine resistant in pancreatic ductal adenocarcinoma. J Pharmacol Sci. 2018;137(2):116–121. doi:10.1016/j.jphs.2018.06.002
20. Li C, Wang F, Wei B, Wang L, Kong D. LncRNA AWPPH promotes osteosarcoma progression via activation of Wnt/beta-catenin pathway through modulating miR-93-3p/FZD7 axis. Biochem Biophys Res Commun. 2019;514(3):1017–1022. doi:10.1016/j.bbrc.2019.04.203
21. Zhang J, Wang Q, Quan Z. Long non-coding RNA CASC9 enhances breast cancer progression by promoting metastasis through the meditation of miR-215/TWIST2 signaling associated with TGF-beta expression. Biochem Biophys Res Commun. 2019;515(4):644–650. doi:10.1016/j.bbrc.2019.05.080
22. Wang J, Zhao X, Guo Z, Ma X, Song Y, Guo Y. Regulation of NEAT1/miR-214-3p on the growth, migration and invasion of endometrial carcinoma cells[J]. Arch Gynecol Obstet. 2017;295(6):1469–1475. doi:10.1007/s00404-017-4365-1
23. Xu C, He T, Li Z, Liu H, Ding B. Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed Pharmacother. 2017;95:1504–1513. doi:10.1016/j.biopha.2017.08.097
24. Zhu WS, Tang CM, Xiao Z, et al. Targeting EZH1 and EZH2 contributes to the suppression of fibrosis-associated genes by miR-214-3p in cardiac myofibroblasts. Oncotarget. 2016;7(48):78331–78342. doi:10.18632/oncotarget.13048
25. Xu Y, Zhang G, Zou C, et al. LncRNA MT1JP suppresses gastric cancer cell proliferation and migration through MT1JP/MiR-214-3p/RUNX3 axis. Cell Physiol Biochem. 2018;46(6):2445–2459. doi:10.1159/000489651
26. Cai H, Miao M, Wang Z. miR-214-3p promotes the proliferation, migration and invasion of osteosarcoma cells by targeting CADM1. Oncol Lett. 2018;16(2):2620–2628. doi:10.3892/ol.2018.8927
27. Perera RM, Stoykova S, Nicolay BN, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524(7565):361–365. doi:10.1038/nature14587
28. Sousa CM, Biancur DE, Wang X, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536(7617):479–483. doi:10.1038/nature19084
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.