Back to Journals » OncoTargets and Therapy » Volume 13
Long Noncoding RNA H19 Facilitates Small Cell Lung Cancer Tumorigenesis Through miR-140-5p/FGF9 Axis
Authors Li X, Lv F, Li F, Du M, Liang Y, Ju S, Liu Z, Wang B, Gao Y
Received 12 January 2020
Accepted for publication 10 April 2020
Published 28 April 2020 Volume 2020:13 Pages 3525—3534
DOI https://doi.org/10.2147/OTT.S245710
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Xingkai Li, Fang Lv, Fang Li, Minjun Du, Yicheng Liang, Shaolong Ju, Zixu Liu, Bing Wang, Yushun Gao
National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, People’s Republic of China
Correspondence: Bing Wang; Yushun Gao Tel +86-13801308915; +86-13801185069
Email [email protected]; [email protected]
Purpose: Long noncoding RNAs (lncRNAs) are emerging as gene regulators to drive many important cancer phenotypes through interaction with microRNAs. There have been numerous data about upregulation of H19 and its strong oncogenic function in progression of cancers. However, the function and detailed mechanisms of H19 on small cell lung cancer (SCLC) are still unclear.
Methods: In this study, we investigated H19 expression in SCLC and para-carcinoma tissues. We also explored the function and detailed mechanisms of H19 on SCLC cells via RT-PCR, transwell assay, Western blot, dual-luciferase report assay and RNA pull-down experiments.
Results: In this study, we observed that H19 was upregulated in SCLC compared with para-carcinoma tissues or NSCLC tissues. We also uncovered that H19 could promote proliferation and migration of SCLC cells. Functional investigation illustrated that H19 acted as a sponge for miR-140-5p to regulate its expression in SCLC. Interestingly, we further found that H19 upregulated FGF9 expression to promote SCLC progression via sponging miR-140-5p. H19 and FGF9 were also revealed to have similar expression patterns in clinical SCLC samples.
Conclusion: These data demonstrated that H19 might be a promising prognostic and therapeutic target for SCLC.
Keywords: H19, small cell lung cancer, SCLC, miR-140-5p, FGF9
Corrigendum for this paper has been published
Introduction
Among all the cancers, lung carcinoma is one of the most frequently diagnosed cancers at the global level, and it has the highest mortality rate, being responsible for 18.4% of all cancer-related deaths worldwide.1–3 Small cell lung cancer (SCLC) possesses about 15% of all lung cancers, is a minor subtype of lung cancer. Due to early development of widespread metastasis, SCLC was considered as an aggressive high-grade neuroendocrine malignancy associated with a poor prognosis.4,5 As a result, treatment options are often not available and limited for SCLC.6 Therefore, it is urgent to explore novel molecules for accurate prognostic prediction and targeted therapy.
More and more long noncoding RNAs (lncRNAs) were identified and investigated with the rapid development of genome sequence, which played indispensable role in the development and progression of cancers.7–9 LncRNA H19 is located at the human chromosome 11p15 near the telomeric region with 2356 nucleotides.10 There have been numerous articles about upregulation of H19 and its strong oncogenic function in cancer progression, such as breast cancer,11 colon cancer,12 prostate cancer,13 esophageal squamous cell cancer,14 multiple myeloma,15 hepatocellular carcinoma,16 non-small cell lung cancer17 and so on. However, the role of H19 in small cell lung cancer is unclear.
In our preliminary experiment, we observed that H19 expression in SCLC tissues was obviously elevated compared with adjacent non-tumor tissues or NSCLC tissues. Mechanically, H19 acted as a sponge for miR-140-5p to increase FGF9 expression, eventually promoting cancer cell proliferation and migration, suggesting that H19 maybe function as an oncogene in the progression of small cell lung cancer.
Materials and Methods
Patient Tissue Samples
In this research, total of 120 clinical tissue specimens (40 SCLC tissues, 40 NSCLC tissues and 40 adjacent normal tissues) were received from Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. Patients who did not receive neoadjuvant radiotherapy or chemotherapy before surgery. Meanwhile, the clinicopathological data were recorded in detail. The study was approved by the ethics committee of National Cancer Center, Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. And all enrolled patients provided written informed consent.
Cell Culture
Human non-small cell lung cancer cell lines (A549, H1299, H460, PC9), small cell lung cancer cell lines (H446, H69, SHP-77, DMS-53) and normal epithelial cell line BEAS-2B were purchased from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). These cells were cultured in DMEM or RPMI1640 medium, containing 10% fetal bovine serum at 37°C with 5% CO2 in a humidified incubator.
RNA Isolation, Reverse Transcription and Quantitative Real-Time PCR
Total RNA from cells or tissues were extracted using TRIzol reagent according to the manufacturer’s instructions. Then, concentration and quality of RNA was measured via Nanodrop. cDNA was gained from RNA using Prime Script RT-PCR kit (Takara, Japan). Lastly, cDNA was used to perform PCR on Real-Time PCR Detection System (Applied Biosystems, Foster City, CA). The primer pairs used for qRT-PCR were as follows:
FGF9 forward primer: 5ʹ- TGGACAGCCCGGTTTTGTTA −3ʹ,
reverse primer: 5ʹ- TCCAGAATGCCAAATCGGCT −3ʹ;
H19 forward primer: 5ʹ- ATCGGTGCCTCAGCGTTCGG −3ʹ,
reverse primer: 5ʹ- TCCTCGCCGTCACACCG −3ʹ;
GAPDH forward primer: 5ʹ- GGTGAAGGTCGGAGTCAACG −3ʹ,
reverse primer: 5ʹ- ACCATGTAGTTGAGGTCAATGA −3ʹ;
MiR-140-5p forward primer: 5ʹ- TGCGGCAGTGGTTTTACCCTATG −3ʹ,
reverse primer: 5ʹ- CCAGTGCAGGGTCCGAGGT −3ʹ;
U6 forward primer: 5ʹ- GCTTCGGCAGCACATATACT-3ʹ,
reverse primer: 5ʹ- CGCTTCACGAATTTGCGTGT −3ʹ;
Overall Survival Analysis
We sort from high to low according to H19 expression, and select the top 50% as the high expression group and the last 50% as the low expression group. After allocating the patients into different groups, Kaplan–Meier survival curves were prepared for each of the patient groups. The clinical data were from 40 SCLC tissues of Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. The statistical significance of the prognosis difference was inferred with the Log rank test.
Vector Construction, si/shRNA Synthesis and Cell Transfection
The cDNA encoding the CDS of H19 was PCR-amplified by Thermo Scientific Phusion Flash High-Fidelity PCR Master Mix and subcloned into the EcoRI and NotI sites of the pcDNA3.1 vector, named pcDNA3.1-H19. The siRNAs and shRNA targeting H19, mi140-5p mimic and inhibitor were purchased from Suzhou Synbio Technologies. SCLC cells were transfected plasmid or siRNA according to appropriate cell density and manufacturer’s instructions. As to transient transfection of siRNA or miRNA, it was performed using lipofectamine RNAiMAX (Invitrogen). Lipofectamine2000 (Invitrogen) was employed with plasmid transfection. The siRNA sequences were as follows:
siH19-1 sense: 5ʹ- ACGAGGCACUGCGGCCCAG −3ʹ;
siH19-2 sense: 5ʹ- GGCCAAGACGCCAGGUCCG −3ʹ;
siFGF9 sense: 5ʹ- CUGGAUUUCACUUAGAAAU −3ʹ;
siNC sense: 5ʹ- ACGUGACACGUUCGGAGAA −3ʹ;
MiR-140-5p mimic sense: 5ʹ- CAGUGGUUUUACCCUAUGGUAG −3ʹ;
MiR-140-5p inhibitor sense: 5ʹ- CUACCAUAGGGUAAAACCACUG −3ʹ;
MiR-NC sense: 5ʹ- UUGUACUACACAAAAGUACUG −3ʹ;
Western Blotting
The protein samples were extracted via RIPA lysis from cells on ice, and the concentration was detected using the bicinchoninic acid (BCA) assay (Pierce). The protein was boiled in 1* SDS loading buffer and loaded into 10% SDS-PAGE with 50 μg in each well and then transferred onto nitrocellulose membranes, which was then blocked in 5% skim milk powder solution. After washing, the primary antibodies against FGF9 (sc-8413, Santa Cruz Biotechnology) or GAPDH (ab181602) were added and incubated overnight at 4°C. Next day, the membranes were incubated with secondary antibody for 1 h at RT after washed with TBST buffer 4 times (5 min each time). Then, they were washed with TBST 4 times (5 min each time). Lastly, signals were quantified and analyzed using the Image-Pro Plus 6.0 software (Media Cybernetics, Sarasota, USA). GAPDH acts as an internal control.
Cell Counting Kit-8
For cancer cell proliferation assay, the cells were seeded into plate and transfected with siRNA, miRNA or plasmid according to appropriate protocol. At each time point, CCK-8 reagent was added into each well. After incubation, OD450 absorbance was measured to reflect cell viability.
Transwell Migration Assay
In vitro cell migration assay was detected using 8um pore-size transwell. 100µl cell suspension (1*104) were seeded into transwell. Then, transwell was put in 24-well plates. 600µl complete medium was added to the bottom wells of the chambers. After 24 h, the transwell was removed and fixed with methanol for 10min, the upper cells were scraped using cotton ball, the cells of bottom surface were stained with crystal violet.
Dual-Luciferase Report Assay
The wide-type or mutation type sequence of H19 was synthesized and cloned into pmirGLO luciferase vector with miR-140-5p. Then, cells were seeded into 24 well dish and co-transfected with relevant plasmid (H19-WT, H19-mut) and miR-140-5p. After incubation for 48 h, the luciferase activity was detected via Dual-luciferase reporter system.
RNA Immunoprecipitation (RIP) Assay
RIP was performed as described.18 The cells were harvested and lysed with lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.05% Igepal, 0.5% NP-40, 0.5 mM PMSF, 2 mM RVC, protease inhibitor cocktail (Roche)), then the supernatant was collected. 10% cell extract was used as Input, and the other part was co‐precipitated with antibodies rabbit anti-AGO2 (ab186733, 5ug) or IgG (ab109489, 5ug). The magnetic beads‐antibody complex was washing with RIP wash buffer (50 mM Tris pH 7.4, 300 mM NaCl, 0.05% Sodium Deoxycholate, 0.5% NP-40, 0.5 mM PMSF, 2 mM RVC, protease inhibitor cocktail (Roche)) for 4 times. Then, it was eluted with 100 µL cell elution buffer. After being detached by protease K, RNA of the samples and input was collected for the subsequent PCR quantification.
RNA Pull‐down
The biotin‐labeled RNA was in combination with Streptavidin C1 Dynabeads (cat: 65,002; invitrogen) according to the manufacturer’s instructions. The cells were harvested and lysed with lysis buffer (Ambion, Austin, TX). The lysate was incubated with bio-RNA-Beads complex overnight at 4°C on a rotating platform. After washing with wash buffer for 4 times, the bound RNA was extracted with Trizol, and H19 enrichment was determined by RT‐qPCR.
Statistical Analysis
The measurement data were expressed as the mean ± standard deviation from at least three independent experiments. All statistical analyses were performed using Graphpad Prism software. Pearson analysis was used to determine the correlation between miR-140-5p and H19 or FGF9; t-test was used for difference comparison between experimental and control groups. P < 0.05 indicative of statistically significant.
Result
LncRNA H19 Was Overexpressed in SCLC Tissues and Cells
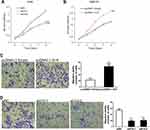
To elucidate the expressive discrepancy of H19 in SCLC tissues, NSCLC tissues and adjacent non-tumor tissues, qRT-PCR was performed from clinical specimens. We observed that H19 expression in SCLC tissues was obviously elevated compared with adjacent non-tumor tissues or NSCLC tissues (Figure 1A). Similarly, the expression of H19 was significantly higher in SCLC cell lines (H446, H69, SHP-77, DMS-53) than human bronchial epithelial cell line BEAS-2B or NSCLC cell lines (A549, H1299, H460, PC9) (Figure 1B). Then, we further investigated the correlations between H19 expression and clinicopathological features in SCLC tissues. Patients were divided into high and low expression groups according to the median level of H19 expression in SCLC tissues. The results illustrated that H19 upregulation was correlated with lymph node metastasis and disease stage, but not associated with patient age or smoking history (Table 1). Moreover, patients with high H19 expression had a significantly poorer prognosis than those with low H19 expression (Figure 1C). From these results above, it is implied that lncRNA H19 increased expression may be important in the progression of SCLC.
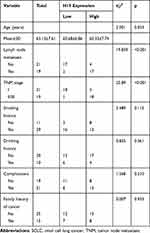 |
Table 1 Correlation Between H19 Expression and Clinicopathological Features in SCLC Patients (n=40) |
H19 Promoted SCLC Cell Proliferation and Migration
To determine whether H19 played a role in SCLC progression, the loss and gain of function study were conducted. We knocked down H19 expression via transfecting siRNA-H19 in H446 cell line and ectopically overexpressed H19 by transfecting pcDNA3.1-H19. We observed that H19 knockdown could inhibit cell proliferation, whereas overexpression of H19 increased this effect by CCK-8 assay (Figure 2A and B). Meanwhile, the shRNA for stable knockdown of H19 was constructed to silence H19 expression level (Figure S1A). Also, the results indicated that H19 stable knockdown decreased cell proliferative ability (Figure S1B). We next examined the function of H19 on cell migration capacity via transwell assay. The result indicated that H19 knockdown conspicuously suppressed SCLC cell migration capacity (Figure 2D, S1C). In contrast, H19 overexpression substantially enhanced SCLC cell migration capacity (Figure 2C). Therefore, these data strongly suggested that H19 played an oncogenic activity in SCLC cell progression.
H19 Directly Bound and Negative Regulated Expression of miR-140-5p
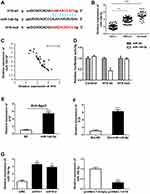
There is growing evidence that lncRNAs can function as ceRNAs by competitively binding miRNAs. We first predicted the potential targets of H19 using Starbase and RNA22 databases. The bioinformatics analysis revealed that H19 contained complementary sequence binding with miR-140-5p (Figure 3A). We found miR-140-5p expression in SCLC tissues was obviously lower than adjacent non-tumor tissues or NSCLC tissues (Figure 3B). To explore the correlation between H19 and miR-140-5p in SCLC tissues, RT-qPCR was performed and showed that H19 expression was inverse associated with the expression level of miR-140-5p (Figure 3C). To further verify whether H19 directly binds to miR-140-5p, we performed a dual-luciferase reporter assay. It showed that luciferase activity of H19-wt was obviously reduced in comparison with H19-mut group (Figure 3D). It is widely known that miRNA can bind Ago2 to posttranscriptionally regulate gene expression. AGO2-RIP assay was performed to determine whether H19 bound to miR-140-5p in this way. The result implied that H19 was enriched in Ago2-RIP in miR-140-5p-transfected cells (Figure 3E). In addition, RNA pull-down assay further showed H19 expression level was largely enriched by bio-miR-140-5p (Figure 3F). Moreover, qRT-PCR results indicated that H19 significantly reduced miR-140-5p expression (Figure 3G). These results suggested that miR-140-5p directly bound to H19 and was negatively regulated by H19.
H19 Promoted SCLC Progression Through Negative Regulation of miR-140-5p
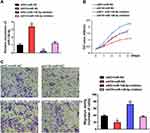
MiR-140-5p has been identified as a tumor suppressor gene in multiple cancer progression. However, the function of miR-140-5p on small cell lung cancer is still unclear. To explore the relationship between H19 and miR-140-5p involving cancer cell proliferation and migration. Rescue experiments were performed through transfecting siRNA H19 and miR-140-5p inhibitor. The expression level of miR-140-5p was reversed by siRNA H19 (Figure 4A). By CCK8 assay, we found that H19 promoted cell proliferation, miR-140-5p inhibited cell proliferation, whereas when cells co-transfected with siRNA H19 and miR-140-5p inhibitor, miR-140-5p inhibitor could reverse the H19 knockdown mediated cell viability (Figure 4B). Similarly, transwell assay indicated that miR-140-5p inhibitor enhanced cell migration capacity reduced by H19 knockdown (Figure 4C). Taken together, these results implied that miR-140-5p inhibition could reverse H19 knockdown induced anti-tumor role in SCLC.
H19 Promoted SCLC Progression Through miR-140-5p/FGF9 Pathway
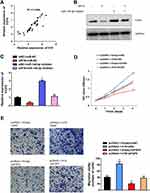
It is reported that miR-140-5p suppressed hepatocellular carcinoma progression by targeting fibroblast growth factor 9 (FGF9).19 Although FGF9 is strongly associated with malignant transformation and has been reported as a notable oncogene in several cancers.20,21 We ask whether H19 expedites SCLC development via targeting miR-140-5p/FGF9 axis. Firstly, we found that mRNA expression pattern of H19 was positive correlated with FGF9 through Spearman correlation analysis (Figure 5A). Then, rescue experiments have shown that H19 knockdown could reverse miR-140-5p inhibitor mediated increase in FGF mRNA and protein expression levels (Figure 5B and C). Furthermore, H19 overexpression could rescue the cell proliferation and migration capability restrained by FGF9 knockdown (Figure 5D and E). Overall, these results demonstrated that H19 promoted SCLC tumorigenesis by sponging miR-140-5p to increase FGF9 expression level.
Discussion
Over the past decade, small cell lung cancer (SCLC) is defined as “refractory cancer” because of its widespread metastasis and poor prognosis, with a 5-year survival rate of only 2–8%.22–24 In this context, unfortunately, SCLC patients are still short of effective therapies. Hence, SCLC urgently needs treatment innovation. Increasing evidence shows that lncRNAs play an important role in the occurrence and development of many cancers, including solid and blood tumors.25–28 However, very few studies have involved small cell lung cancer. For example, Linc00173 promotes chemoresistance of small cell lung cancer by sponging miR-218.29 LncRNA TUG1 is involved in small cell lung cancer progression by regulating LIMK2b expression.30
In this study, we first uncovered the role of H19 in small cell lung cancer progression. H19, a highly conserved lncRNA, is abnormally expressed in human malignant tumors, and regulates the hallmarks of cancer through various molecular mechanisms, thus playing a carcinogenic or anti-cancer role.10,31 However, the role of H19 in small cell lung cancer has not been reported. Our study found that H19 was significantly upregulated in SCLC tissues and cell lines compared to NSCLC tissue and adjacent tissues and cells. Subsequently, we explored the function of H19 on proliferation and migration of SCLC cells. We demonstrated that H19 knockdown significantly inhibited the proliferation and migration ability of SCLC. Combining these results, we proposed that H19 may be an important oncogene marker in the progression of SCLC.
One of the main mechanisms of lncRNAs is that they can regulate the function of corresponding target genes by binding miRNAs, thereby affecting the development of tumors.8,32 We discovered that H19 directly bound and negative regulated expression of miR-140-5p. MiR-140-5p has previously been documented as a tumor suppressor gene in many studies.33,34 Following, we uncovered that FGF9 was a direct target of miR-140-5p in SCLC, which has been reported as a notable oncogene in several cancers. It is worth noting that the expression level of FGF9 is co-regulated by H19 and miR-140-5p in small cell lung cancer cell lines. In addition, the cell phenotype caused by FGF9 silencing was eliminated via H19 overexpression. In summary, all data show that H19 acted as a sponge of miR-140-5p to isolate FGF9 during SCLC progression.
Conclusion
Take together, we reported an important oncogene lncRNA H19 in SCLC progression. Our results indicated that knockdown of H19 could significantly reduce the ability of cell proliferation and metastasis. Moreover, H19 promoted SCLC tumorigenesis through miR-140-5p/FGF9 axis, which further emphasizes the potential of this axis in SCLC diagnosis and therapy. However, the detailed mechanism of H19 in SCLC still needs to be further explored.
Ethics and Consent Statement
Informed consent was obtained from all individual participants included in the study.
All procedures performed in studies involving humans were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
Materials and Correspondence
Correspondence and material requests should be addressed to Yushun Gao.
Acknowledgments
The work was funded by the National key R&D Program of China (Grant Nos. 2016YFC0901401).
Author Contributions
All authors read and approved the final manuscript. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no competing interests.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Jemal A, Miller KD, Ma J, et al. Higher lung cancer incidence in young women than young men in the United States. N Engl J Med. 2018;378(21):1999–2009. doi:10.1056/NEJMoa1715907
3. Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–1953. doi:10.1002/ijc.31937
4. Sabari JK, Lok BH, Laird JH, Poirier JT, Rudin CM. Unravelling the biology of SCLC: implications for therapy. Nat Rev Clin Oncol. 2017;14(9):549–561. doi:10.1038/nrclinonc.2017.71
5. Tsiouprou I, Zaharias A, Spyratos D. The role of immunotherapy in extensive stage small-cell lung cancer: a review of the literature. Can Respir J. 2019;2019:6860432. doi:10.1155/2019/6860432
6. van de Kamp HJ, Molder MT, Schulkes KJG, et al. Impact of lung cancer treatment on cognitive functioning. Clin Lung Cancer. 2020;21(2):114–126.
7. Zhao Y, Li H, Fang S, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44(D1):D203–D208. doi:10.1093/nar/gkv1252
8. Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109(7):2093–2100. doi:10.1111/cas.13642
9. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.010
10. Lecerf C, Le Bourhis X, Adriaenssens E. The long non-coding RNA H19: an active player with multiple facets to sustain the hallmarks of cancer. Cell Mol Life Sci. 2019;76(23):4673–4687. doi:10.1007/s00018-019-03240-z
11. Zhou W, Ye X-L, Xu J, et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10(483):eaak9557. doi:10.1126/scisignal.aak9557
12. Safi R, Mohsen-Kanson T, Nemer G, et al. Loss of ferrochelatase is protective against colon cancer cells: ferrochelatase a possible regulator of the long noncoding RNA H19. J Gastrointest Oncol. 2019;10(5):859–868. doi:10.21037/jgo.2019.03.09
13. Bacci L, Aiello A, Ripoli C, et al. H19-dependent transcriptional regulation of β3 and β4 Integrins upon estrogen and hypoxia favors metastatic potential in prostate cancer. Int J Mol Sci. 2019;20(16):4012. doi:10.3390/ijms20164012
14. Li X, Yang H, Wang J, et al. High level of lncRNA H19 expression is associated with shorter survival in esophageal squamous cell cancer patients. Pathol Res Pract. 2019;215(11):152638. doi:10.1016/j.prp.2019.152638
15. Zheng J-F, Guo N-H, Zi F-M, Cheng J. Long non-coding RNA H19 promotes tumorigenesis of multiple myeloma by activating BRD4 signaling by targeting miR-152-3p. Mol Cell Biol. 2019. doi:10.1128/MCB.00382-19
16. Ding K, Liao Y, Gong D, Zhao X, Ji W. Effect of long non-coding RNA H19 on oxidative stress and chemotherapy resistance of CD133+ cancer stem cells via the MAPK/ERK signaling pathway in hepatocellular carcinoma. Biochem Biophys Res Commun. 2018;502(2):194–201. doi:10.1016/j.bbrc.2018.05.143
17. Zheng ZH, Wu DM, Fan SH, Zhang ZF, Chen GQ, Lu J. Upregulation of miR-675-5p induced by lncRNA H19 was associated with tumor progression and development by targeting tumor suppressor p53 in non-small cell lung cancer. J Cell Biochem. 2019;120(11):18724–18735. doi:10.1002/jcb.29182
18. Xing Y-H, Yao R-W, Zhang Y, et al. SLERT regulates DDX21 rings associated with pol I transcription. Cell. 2017;169(4):664–678.e616. doi:10.1016/j.cell.2017.04.011
19. Yang H, Fang F, Chang R, Yang L. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor β receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58(1):205–217. doi:10.1002/hep.26315
20. Mizukami T, Togashi Y, Naruki S, et al. Significance of FGF9 gene in resistance to anti-EGFR therapies targeting colorectal cancer: a subset of colorectal cancer patients with FGF9 upregulation may be resistant to anti-EGFR therapies. Mol Carcinog. 2017;56(1):106–117. doi:10.1002/mc.22476
21. He X, Chen SY, Yang Z, et al. miR-4317 suppresses non-small cell lung cancer (NSCLC) by targeting fibroblast growth factor 9 (FGF9) and cyclin D2 (CCND2). J Exp Clin Cancer Res. 2018;37(1):230. doi:10.1186/s13046-018-0882-4
22. Gelsomino F, Lamberti G, Parisi C, et al. The evolving landscape of immunotherapy in small-cell lung cancer: a focus on predictive biomarkers. Cancer Treat Rev. 2019;79:101887. doi:10.1016/j.ctrv.2019.08.003
23. Tsoukalas N, Aravantinou-Fatorou E, Baxevanos P, et al. Advanced small cell lung cancer (SCLC): new challenges and new expectations. Ann Transl Med. 2018;6(8):145. doi:10.21037/atm.2018.03.31
24. Zimmerman S, Das A, Wang S, Julian R, Gandhi L, Wolf J. 2017–2018 scientific advances in thoracic oncology: small cell lung cancer. J Thorac Oncol. 2019;14(5):768–783. doi:10.1016/j.jtho.2019.01.022
25. Li X, Wang X, Song W, et al. Oncogenic properties of NEAT1 in prostate cancer cells depend on the CDC5L-AGRN transcriptional regulation circuit. Cancer Res. 2018;78(15):4138–4149. doi:10.1158/0008-5472.CAN-18-0688
26. Chen F, Chen J, Yang L, et al. Extracellular vesicle-packaged HIF-1alpha-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21(4):498–510. doi:10.1038/s41556-019-0299-0
27. Tang Y, Cheung BB, Atmadibrata B, et al. The regulatory role of long noncoding RNAs in cancer. Cancer Lett. 2017;391:12–19. doi:10.1016/j.canlet.2017.01.010
28. Garzon R, Volinia S, Papaioannou D, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2014;111(52):18679–18684. doi:10.1073/pnas.1422050112
29. Zeng F, Wang Q, Wang S, et al. Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate Etk expression. Oncogene. 2020;39(2):293–307.
30. Niu Y, Ma F, Huang W, et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer. 2017;16(1):5. doi:10.1186/s12943-016-0575-6
31. Ye Y, Shen A, Liu A. Long non-coding RNA H19 and cancer: a competing endogenous RNA. Bull Cancer. 2019;106(12):1152–1159. doi:10.1016/j.bulcan.2019.08.011
32. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi:10.1038/nrg.2015.10
33. Zhao J, Bai X, Feng C, Shang X, Xi Y. Long non-coding RNA HCP5 facilitates cell invasion and epithelial-mesenchymal transition in oral squamous cell carcinoma by miR-140-5p/SOX4 axis. Cancer Manag Res. 2019;11:10455–10462. doi:10.2147/CMAR.S230324
34. Fang Z, Yin S, Sun R, et al. miR-140-5p suppresses the proliferation, migration and invasion of gastric cancer by regulating YES1. Mol Cancer. 2017;16(1):139. doi:10.1186/s12943-017-0708-6
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.