Back to Journals » OncoTargets and Therapy » Volume 9
Long noncoding RNA AK126698 inhibits proliferation and migration of non-small cell lung cancer cells by targeting Frizzled-8 and suppressing Wnt/β-catenin signaling pathway
Authors Fu X, Li H, Liu C, Hu B, Li T, Wang Y
Received 16 November 2015
Accepted for publication 29 January 2016
Published 24 June 2016 Volume 2016:9 Pages 3815—3827
DOI https://doi.org/10.2147/OTT.S100633
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Xiao Fu,1 Hui Li,1 Chunxiao Liu,2 Bin Hu,1 Tong Li,1 Yang Wang1
1Department of Thoracic Surgery, Beijing Chao-Yang Hospital, Capital Medical University, Beijing, 2Department of Cardiovascular Surgery, Qilu Hospital of Shandong University, Jinan, Shandong, People’s Republic of China
Background: Recent studies indicate that long noncoding RNAs (lncRNAs) play a key role in the control of cellular processes such as proliferation, metastasis, and differentiation. The lncRNA dysregulation has been identified in all types of cancer. We previously found that lncRNA AK126698 suppresses cisplatin resistance in A549 cells through the Wnt/β-catenin signaling pathway. However, the clinical significance of lncRNA AK126698 and the molecular mechanisms through which it regulates cancer cell proliferation and migration are largely unknown.
Methods: We examined the expression of lncRNA AK126698 in 56 non-small cell lung cancer (NSCLC) tissue samples and three NSCLC cell lines using quantitative real-time polymerase chain reaction. Gain and loss of function approaches were used to evaluate the biological function of AK126698 in NSCLC cells. The effects of lncRNA AK126698 on cell proliferation were investigated using cell counting kit-8 and 5-ethynyl-2'-deoxyuridine assays, and apoptosis was measured by flow cytometry. Protein levels of AK126698 targets were evaluated by Western blotting.
Results: Our results showed that lncRNA AK126698 was significantly downregulated in NSCLC tissues, compared with paired adjacent nontumor tissue samples. Furthermore, lower AK126698 expression was associated with larger tumor size and advanced tumor stage. Ectopic AK126698 expression inhibited cell proliferation and migration and induced apoptosis. Conversely, decreased AK126698 expression promoted cell proliferation and migration and inhibited cell apoptosis. Importantly, we demonstrated that Frizzled-8, a receptor of Wnt/β-catenin pathway, was a target of AK126698. Furthermore, AK126698 could inhibit the activation of Wnt/β-catenin pathway, which was demonstrated by measuring the expression levels of Axin1, β-catenin, c-myc, cyclin D1, and E-cadherin.
Conclusion: It was found in the study that lncRNA AK126698 inhibits the proliferation and migration of NSCLC cells by targeting Frizzled-8 to suppress the Wnt/β-catenin signaling pathway. It may provide a new target for therapeutic intervention in NSCLC.
Keywords: long noncoding RNAs, Frizzled-8, NSCLC, Wnt/β-catenin, proliferation, migration
Introduction
Lung cancer is the most common cause of cancer-related deaths worldwide. Non-small cell lung cancer (NSCLC) accounts for 80%–85% of all lung cancers and is generally diagnosed at an advanced stage.1 Despite considerable progress in treating the disease, the outcome of NSCLC remains unfavorable, with a 5-year overall survival rate of 11%–15%.2 The main reason for the high mortality rate is the sustained proliferation and metastatic potential of tumor cells.3 Lung carcinogenesis is a complicated biological process caused by dysregulated expression of many tumor-related genes.4 Therefore, identifying the molecular mechanisms underlying NSCLC development and progression is essential for improving the diagnosis, prevention, and treatment of this disease.
In the past, research into the mechanisms of tumorigenesis mainly concentrated on protein-coding genes. Recently, transcriptome analyses have unraveled that the major part of the human genome encodes noncoding RNAs (ncRNAs), while only 2% encodes protein.5 The ncRNAs are classified as small ncRNAs (shorter than 200 nucleotides) and long ncRNAs (lncRNAs; >200 nucleotides), which are not translated into proteins.6,7 There is increasing evidence that lncRNAs are involved in many biologic processes, including cell proliferation, cell growth, cell cycle progression, and apoptosis.8 Consequently, aberrant lncRNA expression occurs in diverse human diseases, especially cancer.9–11 Hence, identification of cancer-associated lncRNAs and investigation into their molecular mechanisms and biological functions are important for understanding the molecular biology of cancer development and progression.
Wnt/β-catenin signaling pathway plays a crucial role in regulating multiple aspects of tumor development, including lung cancer.12 When Wnt ligands bind to the seven-pass transmembrane Frizzled (FZD) receptor and its coreceptor, low-density lipoprotein receptor-related proteins 5/6 (LRP5/6)/ROR2/RYK, the Wnt/β-catenin pathway is initiated and Disheveled (DVL) recruits the destruction complex (Axin–APC–GSK3β complex) to the plasma membrane, resulting in β-catenin stabilization and subsequent accumulation in the cytoplasm. Free β-catenin accumulates and is translocated to the nucleus, where it binds to the T-cell factor/lymphoid enhancer factor TCF/LEF to regulate the expression of target genes, such as c-myc, cyclin D1, and E-cadherin.13 Thus, FZD is an essential component of Wnt/β-catenin pathway. FZD expression is reported to be upregulated in some cancer tissues.
In a previous study, we had observed that lncRNA AK126698 was significantly downregulated in A549/DDP cells compared with parental A549 cells. In addition, AK126698 regulated cisplatin resistance in A549 cells through the Wnt/β-catenin signaling pathway.14 However, the clinical importance of lncRNA AK126698 and the molecular mechanisms controlling its effects are unknown. Thus, the present study aimed to investigate AK126698 expression in NSCLC tissues compared with adjacent nontumor tissues and its role in the malignant progression of NSCLC. Furthermore, we had previously predicted that FZD8, the most important Wnt receptor, might be a target of AK126698.14 To test this hypothesis, we investigated the influence of AK126698 on the expression of FZD8 and its downstream genes.
Materials and methods
Cell lines and human samples
Three NSCLC cell lines (A549, NCI-H520, and H1299) and a normal human bronchial epithelial cell line (16HBE) were purchased from the Perking Union Medical College, Beijing, People’s Republic of China. The cells were cultured in Roswell Park Memorial Institute 1640 or Dulbecco’s Modified Eagle’s Medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum, 100 μg/mL penicillin, and 100 μg/mL streptomycin at 37°C in a humidified atmosphere containing 5% CO2. Paired NSCLC and adjacent nontumor tissues were obtained from 56 patients who underwent primary NSCLC resection between May and September 2014 in the Department of Thoracic Surgery at Chao-yang Hospital, Capital Medical University, Beijing, People’s Republic of China. None of the patients had received radiotherapy or chemotherapy prior to surgery. Clinical data, including sex and age of patients, tumor size, histologic tumor type, and tumor–node–metastasis stage, were obtained from surgical and pathological records. Tissue samples were collected at surgery, immediately snap-frozen in liquid nitrogen, and stored at −80°C for RNA extraction. All tissue samples were thoroughly reviewed by two experienced pathologists. Written informed consent was obtained from all patients and the study was approved by the Ethics Committee of Chao-yang Hospital.
RNA extraction and quantitative real-time polymerase chain reaction analysis
Total RNA was purified from each sample using the TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) and treated with DNase I (Thermo Fisher Scientific) according to the manufacturer’s instructions. Total RNA was quantified by measuring the absorbance at 260 and 280 nm. Only samples with an A260:A280 ratio between 1.8 and 2.1 were analyzed further. Complementary DNA was obtained using a PrimeScript RT Reagent Kit with a gDNA Eraser (TaKaRa Biotechnology [Dalian] Co, Ltd., Dalian, People’s Republic of China), and real-time quantitative polymerase chain reaction (qRT-PCR) was performed using TransStart® Tip Green qPCR SuperMix (Transgen, Beijing, People’s Republic of China). The samples were cycled at an initial single cycle of 30 seconds at 94°C, followed by 40 cycles of 5 seconds at 94°C, 15 seconds at 60°C, and 34 seconds at 72°C. The expression of 18S was used as the internal control. Their sequences were as follows:
AK126698 sense 5′-CTGGCAAGTTCTCATCCACA-3′;
reverse 5′-ATGTTGGCCAAGTTGGTCTC-3′;
FZD8 sense 5′-GGACTACAACCGACCT-3′;
reverse 5′-ACCACAGGCCGATCCAGAAGAC-3′;
LRP6 sense 5′-GCTCAGAGTCCCAGTTCCAG-3′;
reverse 5′-TCCCTTCATACGTGGACACA-3′;
18S sense 5′-GTAACCCGTTGAACCCCATT-3′;
reverse 5′-GTAACCCGTTGAACCCCATT-3′.
Data are the mean ± standard deviation of three independent experiments. Relative quantification of AK1266698, FZD8, and LRP6 expression was calculated using the 2−ΔΔCT method relative to 18S.
Cell proliferation assay
Cell proliferation was determined using the cell counting kit-8 (CCK-8) assay kit (Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to the manufacturer’s instructions. Cells were seeded at an initial density of 2×104 cells/mL in 96-well plates. At 12, 24, 48, or 72 hours, CCK-8 (10 μL/well) solution was added to measure cell viability. After 2 hours of incubation at 37°C in 5% CO2, the absorbance of each well was measured at 450 nm.
5-Ethynyl-2′-deoxyuridine incorporation assay
According to the manual of a EdU labeling/detection kit (RiboBio, Guangzhou, People’s Republic of China), 50 μM EdU labeling medium was added to the cell culture to allow incubation for 2 hours at 37°C under 5% CO2. Afterwards, the cells were fixed with 4% paraformaldehyde (pH 7.4) for 30 minutes and incubated with glycine for 5 minutes. After washing with phosphate-buffered saline (PBS), staining with anti-EdU working solution was performed at room temperature for 30 minutes. Following wash with 0.5% Triton X-100 in PBS, the cells were incubated with Hoechst33342 (5 μg/mL) at room temperature for 30 minutes, followed by observation under fluorescent microscopy. The percentage of EdU-positive cells was calculated from five random fields in three wells.
Flow cytometry
Cell apoptosis was quantified using a PE Annexin V Apoptosis Detection Kit II (BD Biosciences, San Jose, CA, USA), which measures the redistribution of phosphatidylserine on the plasma membrane. The cells were washed twice with cold PBS and then resuspended in 1,000 mL binding buffer (10 mM Hepes/NaOH [pH 7.4], 140 mM NaCl, 2.5 mM CaCl2) at a concentration of 1×106 cells/mL. A total of 100 μL of this solution (containing 1×105 cells) was transferred to a 5 mL culture tube, 5 μL Annexin V-PE and 5 μL 7-AAD were added, and cells were gently vortexed. Cells were incubated for 15 minutes at room temperature (25°C) in the dark and then 400 μL binding buffer was added to each tube. The cells were analyzed using flow cytometry (FACScan, BD Biosciences) equipped with CellQuest software (BD Biosciences, San Diego, CA, USA): viable, dead, early apoptotic, and late apoptotic cells could be discriminated. The percentages of early and late apoptotic cells were compared with control groups in each experiment. This assay was repeated three times.
Wound healing assays
The cells were seeded in six-well plates and incubated until 90% confluence in serum-free medium before wounding. A 200 μL tip was used to make a vertical wound and the cells were then washed three times with PBS to remove cell debris. Cell migration into the wounded area was monitored by microscopy at the designated time periods.
Cell migration assay
Cell migration was detected using transwell chambers (8 μm; Corning Incorporated, Corning, NY, USA). A total of 1×105 cells were cultured in 100 μL of serum-free Roswell Park Memorial Institute 1640 medium and added into the upper chamber of 8 μm transwells. The lower chamber was filled with 600 μL of Roswell Park Memorial Institute 1640 medium with 10% fetal bovine serum. After incubating for 48 hours at 37°C, the migrated cells were stained with 0.5% crystal violet solution. The cell numbers were determined by counting the penetrating cells under a microscope at ×200 magnification in random fields in each well. Each experiment was performed in triplicate.
Western blot analysis
Total cell lysates were obtained using radioimmunoprecipitation assay buffer (Solarbio, Beijing, People’s Republic of China) containing 1:100 phenylmethylsulfonyl fluoride and phosphatase inhibitors. Total cellular proteins were extracted and separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Bio-Rad Laboratories Inc., Hercules, CA, USA) and transferred to 0.22 μm nitrocellulose (NC) membranes (Sigma-Aldrich Co., St Louis, MO, USA). The membranes were blocked with 5% nonfat milk at room temperature for 2 hours and then incubated overnight at 4°C with rabbit anti-FZD8 (Sigma-Aldrich Co.), rabbit anti-LRP6 (Santa Cruz Biotechnology Inc., Dallas, TX, USA), rabbit anti-Axin1 (Cell Signaling Technology, Danvers, MA, USA), rabbit anti-c-myc (Cell Signaling Technology), rabbit anti-cyclin D1 (Cell Signaling Technology), rabbit anti-β-catenin (Sigma-Aldrich Co.), rabbit anti-E-cadherin (Cell Signaling Technology), or mouse anti-β-actin antibodies (California Bioscience, Coachella, CA, USA). After incubating with IRDyeTM800 (green)- or IRDyeTM700 (red)-conjugated affinity-purified antirabbit or antimouse IgG (LI-COR, Lincoln, NE, USA), specific bands were visualized. The intensity of protein bands was evaluated using a LI-COR Odyssey infrared double-fluorescence imaging system (LI-COR).
Lentivirus construction and cell transfection
The full-length AK126698 complementary DNA sequence was synthesized based on the AK126698 sequence (NCBI database) and then subcloned into a GV367 vector (GeneChem, Shanghai, People’s Republic of China), and the resulting GV367-AK126698 vector was sequenced. The empty GV367 vector was used as the negative control. To produce infectious viruses, 293T packaging cells were cotransfected with the construction plasmid Helper1.0, the envelope plasmid Helper2.0, and GV367-AK126698 or control lentivirus vector. The cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s protocol. After 48 hours of transfection, the medium was collected, and the virus particles were concentrated by centrifugation and stored at −80°C (2.0×108 titer units).
The A549 and NCI-H520 cells were transduced with the Lv-AK126698 or Lv-AK126698-NC virus (at a multiplicity of infection of 10). The supernatant was removed after 24 hours and fresh culture medium was added to the cells. The infection efficiency was confirmed by fluorescence microscopy and quantitative RT-PCR (qRT-PCR) at 72 hours postinfection, and stable clones were selected after 2 weeks using puromycin (2 μg/mL).
To suppress the lncRNA-AK126698 expression, AK126698 small interfering RNA (siRNA) and negative control siRNA were purchased from RiboBio. Target sequence is sense CUGCAACCCAAGGAAAUAATT, anti-sense UAUUUCCUUGGGUUGCAGTT. The cells were grown on six-well plates to confluency and transfected using riboFECTtm CP (RiboBio, Guangzhou, People’s Republic of China) according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were harvested for qRT-PCR or Western blot analysis.
Statistical analysis
All statistical analyses were performed using Statistical Package for the Social Sciences Version 18.0 software (SPSS Inc., Chicago, IL, USA). Two-tailed Student’s t-test, one-way analysis of variance, and chi-square test were used to analyze the data. The values were expressed as the mean ± standard deviation. Values of P<0.05 were considered statistically significant.
Results
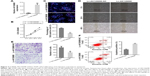
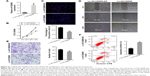
AK126698 expression is frequently downregulated in human NSCLC tissues and cell lines
The level of AK126698 expression was significantly decreased in NSCLC tissues compared with adjacent normal tissues (P<0.05) (Figure 1A). To evaluate the clinical significance of changes in AK126698 expression, we investigated potential correlations between AK126698 expression and clinical parameters. The 56 NSCLC samples were classified into two groups (high and low expression) based on the median level of AK126698 expression. Correlation regression analysis showed that low AK126698 expression correlated with a larger tumor size (P<0.01) (Figure 1B) and advanced pathologic stage (P<0.01) (Figure 1C). In contrast, other clinical parameters such as lymph node metastasis, tumor differentiation, or patient sex did not significantly correlate with AK126698 expression (P>0.05) (Table 1).
  | Table 1 Clinicopathologic association of AK126698 expression in NSCLC |
Next, we examined AK126698 expression in three human NSCLC cell lines (A549, NCI-H520, and H1299) versus a normal human bronchial epithelial cell line (16HBE). AK126698 expression was comparatively low in all three NSCLC cell lines compared with human bronchial epithelial cells (Figure 1D).
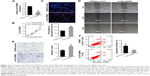
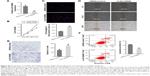
AK126698 overexpression represses cell proliferation and migration and induces apoptosis in NSCLC cells
Based on the significant correlation between low AK126698 expression and large tumor size, we hypothesized that AK126698 has a biological function in NSCLC. To evaluate the biological properties of AK126698 and the effects of AK126698 overexpression on NSCLC, we established A549 cell lines that stably overexpressed AK126698 (Figure 2A). CCK-8 assays showed that AK126698 overexpression significantly repressed cell proliferation in A549 cells compared with controls (Figure 2B). The EdU incorporation assay also indicated that A549 cell growth was significantly inhibited by AK126698 overexpression (Figure 2C). In addition, AK126698 overexpression attenuated migration in Lv-AK126698 cells, as measured by wound healing and transwell migration assays (Figure 2D and E). Apoptosis assays indicated that AK126698 overexpression enhances apoptosis (Figure 2F). Similar results were obtained using the NCI-H520 cells (Figure S1A–F).
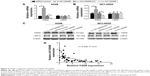
AK126698 knockdown promotes cell proliferation and migration and inhibits apoptosis in NSCLC cells
To further assess the biological function of AK126698 in NSCLC, we inhibited AK126698 expression in NCI-H520 cells by transfection of AK126698-specific siRNA (Figure 3A). CCK-8 assays indicated that AK126698 repression significantly promoted NCI-H520 cell proliferation of cells, compared with controls (Figure 3B). The EdU cell incorporation assay showed that AK126698 knockdown in NCI-H520 cells significantly increased the proportion of EdU-positive cells (Figure 3C). These results suggest that AK126698 repression promotes cell proliferation. To explore the effect of AK126698 downregulation on tumor cell migration, we performed wound healing and transwell assays in NCI-H520 cells after transfection of AK126698-specific siRNA. We found that the siRNA transfection-mediated AK126698 knockdown increased cell migration compared with negative control cells in NCI-H520 (Figure 3D and E). As shown in Figure 3F, siRNA-AK126698 transfection decreased the amount of apoptosis in NCI-H520 cells. Similar results were observed in the A549 cells (Figure S2A–F).
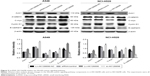
FZD8 is negatively regulated by AK126698
Our previous studies suggested that FZD8 may be a target of AK126698.14 Furthermore, in the current study, we demonstrated that AK126698 overexpression suppressed lung cancer cell proliferation and migration. To understand the molecular mechanism through which AK126698 inhibits NSCLC cell proliferation and migration, we examined whether FZD8 is a downstream effector that mediates the AK126698 function in NSCLC. As shown in Figure 4, AK126698 upregulation significantly inhibited FZD8 mRNA and protein levels in A549 and NCI-H520 cells. Moreover, AK126698 knockdown increased FZD8 mRNA and protein levels in A549 and NCI-H520 cells (Figure 4A–C). In contrast, changes in AK126698 expression level had no effect on LRP6 expression (Figure 4A–C).
As AK126698 negatively regulates FZD8 expression in vitro, we next investigated whether AK126698 modulates FZD8 expression in clinical NSCLC tissues. FZD8 mRNA expression was measured by qRT-PCR in NSCLC samples from 56 patients. Spearman’s rank test found a significant negative correlation between AK126698 and FZD8 expression (r=−0.545, P<0.001) (Figure 4D), supporting a role for AK126698 in regulating FZD8 expression.
AK126698 inhibits Wnt/β-catenin signaling by suppressing FZD8
Wnt/β-catenin signaling plays an important role in NSCLC.12 FZD8 can positively regulate Wnt/β-catenin signaling in NSCLC cells.12 To test whether AK126698-mediated FZD8 downregulation affects Wnt/β-catenin signaling, we measured the levels of Axin1, β-catenin, c-myc, and cyclin D1 proteins, which are important components of the Wnt/β-catenin pathway, and E-cadherin, an important marker of epithelial–mesenchymal transition (EMT) that is controlled by the Wnt/β-catenin pathway. We found that AK126698 overexpression downregulated FZD8, which in turn strongly decreased β-catenin expression and had weaker effects on c-myc and cyclin D1 expression. Conversely, Axin1 and E-cadherin protein levels were increased (Figure 5). Meanwhile, AK126698 knockdown upregulated FZD8, increased β-catenin expression, and enhanced c-myc and cyclin D1 expression, while decreasing Axin1 and E-cadherin protein levels (Figure 5). Together, these data indicate that the AK126698-mediated decrease in cell proliferation and migration may be largely attributed to its ability to inhibit FZD8, which suppresses the Wnt/β-catenin signaling pathway.
Discussion
Developing effective treatments for human cancer is challenging because of the unlimited proliferation, metastasis, and defective apoptosis of cancer cells. Recent studies have revealed that a large number of lncRNAs are involved in the development of human disease, especially cancer.15–17 However, for most lncRNAs, the mechanism of action, biological function, and signaling pathways through which they exert their biological activity remain unknown. The present study aimed to investigate a possible relationship between NSCLC and AK126698, and explore the underlying mechanisms of AK126698.
We found that AK126698 was downregulated in NSCLC tissues compared with adjacent normal lung tissues. More importantly, the level of AK126698 expression correlated with NSCLC tumor size and clinical stage. This is the first report of a relationship between AK126698 expression and NSCLC. This finding is consistent with our observations that AK126698 is downregulated in NSCLC cell lines compared with normal human bronchial epithelial cell line (16HBE). A previous study had reported that AK126698 expression is low in cisplatin-resistant A549/DDP cells compared with parental A549 cells.14 Taken together, these observations indicate that AK126698 may function as a tumor suppressor in NSCLC progression. However, because of the limited follow-up time, it is unknown whether AK126698 expression levels are associated with overall survival in patients with NSCLC.
As low AK126698 expression was associated with a large tumor size and advanced pathologic stage, we speculated that AK126698 could play an important role in tumor cell proliferation and migration. We demonstrated that AK126698 overexpression in A549 and NCI-H520 cells suppressed cell proliferation and migration, whereas AK126698 knockdown enhanced cell proliferation and migration in A549 and NCI-H520 cells. In addition, AK126698 upregulation resulted in increased apoptosis. These results reveal that AK126698-dependent regulation of NSCLC proliferation might involve altering the apoptotic rate.
Aberrant activation of the Wnt/β-catenin signaling pathway has been demonstrated in several cancers, including lung cancer.18,19 Numerous lncRNAs have been demonstrated to target the key components of the Wnt/β-catenin pathway and, thus, modulate cancer cell function. For example, the lncRNA CCAL was found to promote colorectal cancer progression by targeting activator protein 2α and, thus, activating the Wnt/β-catenin pathway.20 Furthermore, HOTAIR inhibits WIF-1 expression, which in turn activates the Wnt/β-catenin pathway.21 Previous studies showed that several FZD receptors including FZD2, FZD4, FZD7, and FZD10 were dysregulated in different types of cancers.22–26 FZD8 upregulation has been identified in lung cancer tissues and cell lines,27,28 and knockdown of FZD8 expression using siRNA has been observed to inhibit the proliferation of tumor cells in vitro and in a xenograft mouse model.28 Therefore, targeted inhibition of FZD8 displays a promising and rational new approach for cancer therapy. We investigated the mechanisms by which AK126698 exerts its function and modulates malignant NSCLC phenotypes. We found that the ability of AK126698 to suppress proliferation and migration is, in large part, owing to its ability to inhabit FZD8 and subsequently activate the Wnt/β-catenin signaling pathway. Furthermore, the coexpression of AK126698 and its target gene FZD8 was detected in NSCLC tissues. We observed that AK126698 was negatively correlated with FZD8 expression in NSCLC tissues. LRP6 is one of the coreceptors of the Wnt/β-catenin pathway and is essential for the activation of the Wnt/β-catenin signaling pathway.29 However, the result of our research indicated that LRP6 was not regulated by AK126698. Taken together, our results strongly suggested that AK126698 suppressed the activation of Wnt/β-catenin pathway by targeting FZD8 in NSCLC development.
Metastasis is the leading cause of death among patients with cancer.30,31 It was found in our study through wound healing and transwell analyses that upregulation of AK126698 expression dramatically weakened the migration ability of NSCLC cells. The Wnt/β-catenin signaling is activated upon the binding of Wnt ligands to FZD/LRP coreceptor complexes, β-catenin escapes from degradation in the Axin complex,32 and accumulated β-catenin is translocated to the nucleus. β-catenin can also activate the expression of Snail and Slug, which represses E-cadherin expression and induces EMT and cancer metastasis.33,34 Recent studies are beginning to reveal the association of lncRNA with EMT.33,35 Increased MALAT-1 led to bladder cancer cell migration by Wnt signaling activation and in turn induced EMT.33 H19 is associated with enhancer of Enhancer of zeste homolog 2 (EZH2), and this association contributes to activate Wnt/β-catenin and subsequent downregulation of E-cadherin.35 The present study also unraveled that upregulated AK126698 contributed to a decreased expression of FZD8, and that this association leads to Wnt/β-catenin inactivation and subsequent upregulation of E-cadherin. The downregulation of AK126698 had the opposite effects.
To summarize, our present study shows that AK126698 is frequently downregulated in NSCLC patients. In addition, AK126698 can significantly inhibit NSCLC cell proliferation and migration and increase apoptosis. Finally, our findings suggest that AK126698 is a negative regulator for the malignant transformation pathway Wnt/β-catenin through targeting FZD8. Our studies suggest that targeting the AK126698/FZD8 interaction or perturbing AK126698 expression is a new potential therapeutic strategy in the treatment of NSCLC.
Acknowledgment
The abstract of this paper was presented at the 16th IASLC World Conference on Lung Cancer with the title, “The Long Non-Coding RNA AK126698 Modulates Wnt/β-catenin Signaling through FZD-8 Silencing in Non-Small-Cell Lung Cancer,” as a poster presentation with interim findings.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. | ||
Raungrut P, Wongkotsila A, Lirdprapamongkol K, et al. Prognostic significance of 14-3-3γ overexpression in advanced non-small cell lung cancer. Asian Pac J Cancer Prev. 2014;15(8):3513–3518. | ||
Koudelakova V, Kneblova M, Trojanec R, Drabek J, Hajduch M. Non-small cell lung cancer – genetic predictors. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2013;157(2):125–136. | ||
Minna JD1, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1(1):49–52. | ||
Hattori M. Finishing the euchromatic sequence of the human genome. Tanpakushitsu Kakusan Koso. 2005;50(2):162–168. | ||
Amaral PP1, Dinger ME, Mercer TR, et al. The eukaryotic genome as an RNA machine. Science. 2008;319(5871):1787–1789. | ||
Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. | ||
Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. | ||
Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. | ||
Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. | ||
Braconi C, Kogure T, Valeri N, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30(47):4750–4756. | ||
Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106(1):djt356. | ||
Ring A, Kim YM, Kahn M. Wnt/catenin signaling in adult stem cell physiology and disease. Stem Cell Rev. 2014;10(4):512–525. | ||
Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8(5):e65309. | ||
Perez DS, Hoage TR, Pritchett JR, et al. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet. 2008;17(5):642–655. | ||
Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. | ||
Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19(R2):R152–R161. | ||
Klaus A1, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8(5):387–398. | ||
Yang F, Bi J, Xue X, et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279(17):3159–3165. | ||
Ma Y, Yang Y, Wang F, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wntβ-catenin signalling pathway via suppression of activator protein 2α. Gut. Epub 2015 May 20. | ||
Ge XS, Ma HJ, Zheng XH, et al. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104(12):1675–1682. | ||
Tanaka S1, Akiyoshi T, Mori M, Wands JR, Sugimachi K. A novel frizzled gene identified in human esophageal carcinoma mediates APC/beta-catenin signals. Proc Natl Acad Sci. 1998;95(17):10164–10169. | ||
Jin X, Jeon HY, Joo KM, et al. Frizzled 4 regulates stemness and invasiveness of migrating glioma cells established by serial intracranial transplantation. Cancer Res. 2011;71(8):3066–3075. | ||
Ueno K, Hazama S, Mitomori S, et al. Down-regulation of frizzled-7 expression decreases survival, invasion and metastatic capabilities of colon cancer cells. Br J Cancer. 2009;101(8):1374–1381. | ||
Fukukawa C, Nagayama S, Tsunoda T, Toguchida J, Nakamura Y, Katagiri T. Activation of the non-canonical Dvl-Rac1-JNK pathway by Frizzled homologue 10 in human synovial sarcoma. Oncogene. 2009;28(8):1110–1120. | ||
Zeng G, Germinaro M, Micsenyi A, et al. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia. 2006;8(4):279–289. | ||
Bravo DT1, Yang YL, Kuchenbecker K, et al. You L. Frizzled-8 receptor is activated by the Wnt-2 ligand in non-small cell lung cancer. BMC Cancer. 2013;13:316. | ||
Wang HQ, Xu ML, Ma J, Zhang Y, Xie CH. Frizzled-8 as a putative therapeutic target in human lung cancer. Biochem Biophys Res Commun. 2012;417(1):62–66. | ||
MacDonald BT, Yokota C, Tamai K, Zeng X, He X. Wnt signal amplification via activity, cooperativity, and regulation of multiple intracellular PPPSP motifs in the Wnt co-receptor LRP6. J Biol Chem. 2008;283(23):16115–16123. | ||
Yu M, Bardia A, Wittner BS, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–584. | ||
Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19(11):1438–1449. | ||
Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3 beta and beta-catenin and promotes GSK-3 beta-dependent phosphorylation of beta-catenin. EMBO J. 1998;17(5):1371–1384. | ||
Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8(9):2289–2294. | ||
Eger A1, Stockinger A, Schaffhauser B, Beug H, Foisner R. Epithelial mesenchymal transition by c-Fos estrogen receptor activation involves nuclear translocation of beta-catenin and upregulation of beta-catenin/lymphoid enhancer binding factor-1 transcriptional activity. J Cell Biol. 2000;148(1):173–188. | ||
Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333(2):213–221. |
Supplementary materials
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.