Back to Journals » International Journal of General Medicine » Volume 15
Long Non-Coding RNAs ANRIL and HOTAIR Upregulation is Associated with Survival in Neonates with Sepsis in a Neonatal Intensive Care Unit
Authors AbdAllah NB, Al Ageeli E , Shbeer A, Abdulhakim JA , Toraih EA , Salman DO, Fawzy MS , Nassar SS
Received 12 May 2022
Accepted for publication 12 July 2022
Published 20 July 2022 Volume 2022:15 Pages 6237—6247
DOI https://doi.org/10.2147/IJGM.S373434
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Nouran B AbdAllah,1 Essam Al Ageeli,2 Abdullah Shbeer,3 Jawaher A Abdulhakim,4 Eman A Toraih,5,6 Doaa O Salman,6 Manal S Fawzy,7,8 Sanaa S Nassar1
1Department of Pediatrics, Faculty of Medicine, Suez Canal University, Ismailia, Egypt; 2Department of Clinical Biochemistry (Medical Genetics), Faculty of Medicine, Jazan University, Jazan, Saudi Arabia; 3Anesthesiology and Intensive Care, Department of Surgery, Faculty of Medicine, Jazan University, Jazan, Saudi Arabia; 4Medical Laboratory Department, College of Applied Medical Sciences, Taibah University, Yanbu, Saudi Arabia; 5Division of Endocrine and Oncologic Surgery, Department of Surgery, School of Medicine, Tulane University, New Orleans, LA, USA; 6Genetics Unit, Department of Histology and Cell Biology, Faculty of Medicine, Suez Canal University, Ismailia, Egypt; 7Department of Medical Biochemistry and Molecular Biology, Faculty of Medicine, Suez Canal University, Ismailia, Egypt; 8Department of Biochemistry, Faculty of Medicine, Northern Border University, Arar, Saudi Arabia
Correspondence: Manal S Fawzy, Department of Medical Biochemistry and Molecular Biology, Faculty of Medicine, Suez Canal University, Ismailia, Egypt, Email [email protected] Eman A Toraih, Division of Endocrine and Oncologic Surgery, Department of Surgery, School of Medicine, Tulane University, New Orleans, LA, USA, Email [email protected]
Background: Recently, long non-coding RNAs (lncRNAs) have emerged as potential molecular biomarkers for sepsis. We aimed to profile the expression signature of three inflammation-related lncRNAs, MALAT1, ANRIL, and HHOTAIR, in the plasma of neonates with sepsis and correlate these signatures with the phenotype.
Patients and Methods: This case–control study included 124 neonates with sepsis (88 survivors/36 non-survivors) admitted to the neonatal ICU and 17 healthy neonates. The relative expressions were quantified by real-time PCR and correlated to the clinic-laboratory data.
Results: The three circulating lncRNAs were upregulated in the cases; the median levels were MALAT1 (median = 1.71, IQR: − 0.5 to 3.27), ANRIL (median = 1.09, IQR: 0.89 to 1.30), and HOTAIR (median = 1.83, IQR: 1.44 to 2.41). Co-expression analysis showed that the three studied lncRNAs were directly correlated (all p-values < 0.001). Overall and stratification by sex analyses revealed significantly higher levels of the three lncRNAs in non-survivors compared to the survivor group (all p-values < 0.001). Principal component analysis showed a clear demarcation between the two study cohorts in males and females. Cohorts with upregulated ANRIL (hazard ratio; HR = 4.21, 95% CI = 1.15– 10.4, p=0.030) and HOTAIR (HR = 2.49, 95% CI = 1.02– 6.05, p=0.044) were at a higher risk of mortality.
Conclusion: Circulatory MALAT1, ANRIL, and HOTAIR were upregulated in neonatal sepsis, and the latter two may have the potential as prognostic biomarkers for survival in neonatal sepsis.
Keywords: neonatal sepsis, long non-coding RNAs, MALAT1, ANRIL, HOTAIR, survival
Introduction
More than 47% of less than five-year-old deaths globally occur in the neonatal period, resulting in 2.4 million deaths yearly.1 Most of these deaths usually occur in low-income countries, and almost one million deaths are attributed to infectious causes, including neonatal sepsis, meningitis, and pneumonia.1,2 On the other hand, the survivors of neonatal sepsis are vulnerable to short- and long-term neurodevelopmental morbidity.3
Pathophysiology of neonatal sepsis relies on the innate immunity of neonates as their adaptive immune response is not fully developed. A more rigorous evaluation of the possible association between genetic signatures and neonatal sepsis is critical. The related neonatal innate and adaptive immune responses showed impairment of some aspects of innate immunity to bacterial infection, particularly in low-birth-weight infants.4–6
The recently recognized family of non-coding RNAs, long non-coding RNAs (lncRNAs; >200 nucleotides), has been implicated in several biological processes, including innate immunity.7 The lncRNA “Metastasis Associated Lung Adenocarcinoma Transcript 1” (MALAT1) upregulation has demonstrated an independent predictive value as a biomarker for diagnosis, severity, and poor prognosis in adults with sepsis.8,9 Also, the antisense non-coding RNA in the INK4 locus (ANRIL) was implicated in regulating inflammation in rats with uric acid nephropathy,10 and the pro-inflammatory genes, including IL6 and IL8, in human endothelial cells under the control of the nuclear factor-κB.11 Gui et al have identified its clinical utility as a biomarker of severity, inflammation, and prognosis in adult sepsis patients.12 Lastly, the lncRNA “HOX Transcript Antisense RNA” (HOTAIR) was observed to promote sepsis progression by regulating interleukin-6 receptor expression via microRNA-211 in a septic rat model.13 Additionally, it was reported to play a vital role in “sepsis-induced acute kidney injury” via regulating apoptosis.14,15
Although dysregulated expression of the lncRNAs mentioned above was associated with sepsis etiopathology in adults, no studies have explored their potential clinical utility as diagnostic and/or prognostic biomarkers in neonatal sepsis.16 To this end, the authors were inspired to explore the potential clinical utility of the lncRNAs MALAT1, ANRIL, and HOTAIR in a sample of neonatal sepsis to confirm their rules in this devastating disorder. The results of this work could help in risk stratification for this vulnerable group of population and provide preliminary data for future molecular targeted therapy.
Materials and Methods
Study Subjects
A total of 124 neonates with sepsis (93 early-onset and 31 late-onset sepsis) and 17 healthy controls were enrolled in the current study after taking informed consent from all their parents or legal guardians. Cohorts were recruited from Suez Canal University Hospital, Egypt, between December 2018 and November 2019. Age enrollment starts from the day of life (DOL) 1 at delivery till DOL 7 of suspected neonates with any manifestations that enable physicians to diagnose neonatal sepsis. Neonates ≥28 weeks gestational age confirmed by “New Ballard Score”,17 of both sex diagnosed with sepsis (defined as bacteremia presented clinically with dysregulation of response to infection in the first four weeks of life),18 or suspected neonatal sepsis (defined as neonates who are delivered to a mother with risk factors for sepsis, including premature rupture of membrane (PROM) >18 hours, urinary tract infection, genital infections, fever, leukocytosis or chorioamnionitis),19 were enrolled. In addition, neonates presented with clinical indicators (ie, observations and events in the baby) of possible/suspected neonatal sepsis/infection, including red flag clinical indicators and other “National Institute for Health and Care Excellence (NICE)” guidelines-related clinical indicators, were recruited.20 Exclusion criteria included newborns with gestational age <28 weeks, gross congenital malformation/genetic syndromes, history of perinatal hypoxia, hypoxic-ischemic encephalopathy, maternal drug abuse, or maternal viral hepatitis. The study was executed following the “Declaration of Helsinki” guidelines and approved by the Ethics Committee of the Faculty of Medicine, Suez Canal University (approval no. 4463).
Clinical Assessment
Early clinical diagnosis of neonatal sepsis is still highly suspected due to the lack of specific signs and symptoms as they are shared with various other neonatal diseases. Blood culture is the gold standard of lab diagnosis but has certain limitations, which delay early identification. Routinely, 7–13% of neonates are treated empirically for suspected sepsis because of these diagnostic limitations. In this sense, we included all neonates with sepsis, either diagnosed with +ve blood cultures or –ve results.6,21
Prenatal maternal history was examined for maternal risk factors such as premature rupture of membrane, maternal diabetes, anemia, hypertension, pre-eclampsia, urinary tract infection, or triple I (intrauterine inflammation, infection, or both). Detailed obstetric history on the mode of delivery, weight at birth, antepartum hemorrhage, maternal antibiotics, or intrapartum fever were obtained. Putative neonatal risk factors were evaluated as “preterm, intrauterine growth retardation, receiving total parental nutrition, mechanical ventilation, umbilical venous catheterization” and other invasive procedures such as a urinary catheter, chest tubes, or long lines. Symptoms in neonatal sepsis are unspecific, as many non-infected neonates display similar symptoms. There is no single-point assessment of clinical signs to diagnose sepsis; hence, our clinical examination included the following: (1) gestational age assessment, weight, and sex of the full-term neonates, (2) general and systemic examination, including (a) the respiratory system: tachypnea, apnea, increased ventilator support, and oxygen desaturation, (b) the cardiovascular system: bradycardia, pallor, hypotension, persistent pulmonary hypertension (PPHN) and decreased perfusion, (c) metabolic changes: hypothermia, hyperthermia, glucose instability, metabolic acidosis, (d) gastrointestinal system: poor feeding, vomiting, diarrhea, jaundice, feeding intolerance, abdominal distension, and ileus (e) neurologic changes: lethargy, hypotonia, decreased activity, and seizures, (f) hematological: disseminated intravascular coagulopathy, purpura, petechiae, and bleeding.
Blood Sample Collection
Qualified nurses collected 5 milliliters of blood on several occasions (ie, not at one time) from a peripheral vein under an aseptic condition from the enrolled neonates upon admission for routine hematological, chemistry, immunological assessment, blood culture, and genetic analysis. Tubes for genetic analysis (1 mL on EDTA tubes) were transferred immediately to the genetic lab within 20 min to be centrifuged with separation of the buffy coat in sterile Eppendorf for the subsequent genetic analysis. Post-treatment samples were withdrawn three days after starting antibiotics (as post-treatment samples) for 43 neonates (only the available samples for comparison).
Blood Culture
As blood culture is the gold standard to define neonatal sepsis, a blood culture sample (0.5–1 mL) taken from a normally sterile site under aseptic measures was taken in pediatric oxoid blood culture bottles and sent to the microbiology lab for cultivation. Blood culture time to positivity (TTP) or turn-around time technique was used. The blood cultures were incubated aerobically at 37°C and observed daily for the first three days to identify any visible microbial growth (turbidity). A preliminary report of findings is available within 48 hours. However, even in the absence of turbidity, the blood was subcultured up to the seventh day, and the report depends on the growth result rather than the observation of the bottles. The final report is released after 5–7 days accordingly.
RNA Extraction and Reverse Transcription (RT)
Extraction of total RNA from the buffy coat separated from the whole blood was carried out using PureLink RNA Mini Kit (Qiagen, Catalog no. 217184) according to the manufacturer’s instructions. RNA concentration and purity were assessed via NanoDrop ND-1000 spectrophotometer (NanoDrop Tech., Inc. Wilmington, DE, USA). Samples with a 260/280 nm absorbance ratio <1.8 were excluded. RNA was converted to complementary/copy DNA (cDNA) using the “high-capacity cDNA Reverse Transcription (RT)” Kit (Applied Biosystems, USA), as described in our previous work.22 The RT reactions were carried out in a “Mastercycler Gradient Thermocycler (Eppendorf, Hamburg, Germany)” at 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min, then held at 4°C. No template and no enzyme samples as negative controls were included in each run. Gene expression of MALAT1, ANRIL, and HOTAIR compared to GAPDH was performed using quantitative real-time polymerase chain reaction (qPCR) following the “Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE)” guidelines.23 The PCR reactions were carried out in duplicate with a final volume of 20 µL, including 1 µL TaqMan® assays (Hs00273907_s1, Hs03300540_m1, Hs05502358_s1, and Hs02786624_g1) that include the pre-designed primer and probe set (Thermo Fisher Scientific) for MALAT1, ANRIL, HOTAIR, and GAPDH, respectively, diluted in RNAse-free water, 1.33 µL RT product, and 2× TaqMan® Universal PCR Master Mix (Applied Biosystems, Waltham, MA, USA) following the previously described protocols.24 The fold change of the studied lncRNA expressions in each case with sepsis relative to the healthy controls was calculated via the Livak and Schmittgen method based on the quantitative cycle (Cq) value (2−ΔΔCq); where “ΔΔCq = (Cq lncRNA – Cq GAPDH) sepsis cases − (Cq lncRNA – Cq GAPDH) healthy neonates”.25
Statistical Analysis
Statistical Package for the Social Sciences (SPSS) for Windows software (version 27.0; IBM SPSS Statistics, USA) and GraphPad Prism 9.1.2 software were used for data analysis. Data were presented as the median and interquartile range or frequency and percentage. Chi-square (χ2), Fisher’s exact, and Mann–Whitney U (MW) tests were used. Spearman correlation analysis was carried out to assess lncRNAs co-expression. Cox proportional regression analysis was performed to identify independent predictor risk factors for mortality. Hazard ratio (HR) and 95% confidence interval (CI) were reported. Statistical significance was set at a p-value <0.05. Heatmap, hierarchical clustering, box plots, and correlation matrix were generated using “reshape2”, “scales”, “RColorBrewer”, “gplots”, “psych”, “factoextra”, “FactoMineR”, and “ggpubr” R package.
Results
Characteristics of the Study Population
The current study included 124 neonates with sepsis (93 early-onset and 31 late-onset sepsis). Two-thirds of them (65.3%) were males, and in-hospital mortality was reported in 29%. Table 1 demonstrates a comparison between 88 survivors of neonates with sepsis and 36 non-survivors. The non-survivors were more likely to be delivered by cesarean section (66.7% vs 46.6%, p=0.049), preterm (63.9% vs 34.1, p=0.002), with inserted umbilical venous catheter (44.4% vs 9.1%, p<0.001). Higher prevalence of deceased neonates presented with hypoxia (86.1% vs 65.9%, p=0.027) and respiratory distress syndrome (55.6% vs 30.7%, p=0.014). Mothers were more likely to have premature rupture of membranes (52.8% vs 28.4%, p=0.010), pre-eclampsia (13.9% vs 0%, p<0.001), and received amoxicillin (27.8% vs 8.0%, p=0.003). Higher frequency of non-survivors’ neonates required ventilation (63.9% vs 29.5%, p<0.001) and surfactant therapy (33.3% vs 10.2%, p=0.001). Non-survivors’ neonates were more likely to develop complications (75% vs 13.6%, p<0.001), such as pulmonary hemorrhage (16.7% vs 0%, p<0.001), air leak syndrome and pneumothorax (19.4% vs 3.4%, p=0.002), respiratory failure (22.2% vs 1.1%, p<0.001), necrotizing enterocolitis (11.1% vs 0%, p=0.002), and multiple organ failure (22.2% vs 0%, p<0.001).
 |
Table 1 Characteristics of Sepsis Patients According to Survival |
The Circulatory Expression Level of Long Non-Coding RNAs
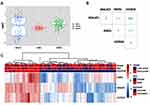
Compared to normal neonates, the three tested lncRNAs expression signatures were upregulated in the circulation of neonates presented with sepsis. Their median levels were as follows: median = 1.71, IQR: −0.5 to 3.27 for MALAT1, median = 1.09, IQR: 0.89 to 1.30 for ANRIL, and median = 1.83, IQR: 1.44 to 2.41 for HOTAIR (Figure 1A). Co-expression analysis showed a direct correlation between the three lncRNAs ranged from weak (r = 0.37) to moderate (r = 0.42 and 0.61) correlations (all p-values <0.001) (Figure 1B). Hierarchical cluster analysis classified patients into two clusters: the first cluster was characterized by high expression of all lncRNAs, and almost all patients died, while the second cluster, including those who survived, exhibited downregulation of at least one of the markers (Figure 1C). On comparison of gene expression levels of the studied lncRNAs pre- and post (three days)-treatment after initiating the antibiotic therapy, there was no observed significant change in their expression signature (Figure S1).
Association of lncRNA Expression with Clinical Features and Survival
The transcriptomic pattern of MALAT1 (p=0.93), ANRIL (p=0.69), and HOTAIR (p=0.98) did not show a significant difference between early-onset and late-onset sepsis groups. However, overall and stratification by sex revealed significantly higher levels of the three lncRNAs in deceased neonates compared to the survivor group (all p-values <0.001) (Figure 2A–C). Analysis of the association of the three studied lncRNAs with other comorbidities in the study groups is presented in Table S1. Running a principal component analysis showed a clear demarcation between the two groups of cohorts in males and females (Figure 3A).
Predictor Risk Factors for Mortality
As depicted in Figure 3B, Cox regression analysis revealed that non-survivor neonates were more likely to be male (HR = 6.43, 95% CI = 1.31–15.7, p = 0.022) and presented with early-onset sepsis (HR = 4.18, 95% CI = 1.05–11.6, p = 0.043). Cohorts with upregulated ANRIL (HR = 4.21, 95% CI = 1.15–10.4, p=0.030) and HOTAIR (HR = 2.49, 95% CI = 1.02–6.05, p = 0.044) were at higher risk of mortality. On the contrary, higher gestational age, more than 33.5 weeks, were less likely to die (HR = 0.79, 95% CI = 0.64–0.96, p=0.020), and those who received surfactant therapy conferred protection (HR = 0.21, 95% CI = 0.06–0.76, p=0.017).
Discussion
Neonatal sepsis contributes to global neonatal morbidity and mortality worldwide, with a higher burden, in particular, in low- and middle-income countries.26 The current study included 124 neonates with sepsis, two-thirds of them were males, and the non-survivor neonates were more likely to be male, as revealed by Cox regression analysis. This male predominance aligns with previous studies that revealed that females have a less exaggerated immune reaction to pathogens than males due to hormonal and genetic/epigenetic modifiers contributing to the observed immunological and survival rate differences.27,28 Also, Cox regression analysis revealed that non-survivor neonates were more likely to be presented with early-onset sepsis. This finding is congruent with recent Fleischmann et al’s meta-nalysis conclusion that “in the overall time frame, estimated incidence and mortality was higher in early-onset than late-onset neonatal sepsis cases” based on 26 studies from fourteen countries.26
In comparison between 88 survivors of neonates with sepsis and 36 non-survivors, the non-survivors were more likely to be delivered by cesarean section, preterm, and have a history of an inserted umbilical venous catheter. Their mothers were more likely to have premature rupture of membranes, pre-eclampsia, and received amoxicillin. Also, non-survivor neonates were more likely to develop complications such as pulmonary hemorrhage, air leak syndrome and pneumothorax, respiratory failure, necrotizing enterocolitis, and multiple organ failure as expected.
On exploring the association of the tested inflammation-related lncRNAs expression signature with neonatal sepsis and outcome, our results demonstrated that the three lncRNAs, MALAT1, ANRIL, and HOTAIR, were markedly increased in the plasma of cases with neonatal sepsis compared to healthy neonates. On correlating the expression signatures of the studied lncRNAs to the clinical features and outcome of the neonatal cases, we found significantly higher levels of the three lncRNAs in deceased neonates than in the survivor group. Furthermore, the cohorts with upregulated ANRIL and HOTAIR were at higher risk of mortality, as revealed by Cox regression analysis.
Given the high blood stability and detection sensitivity of lncRNAs in plasma compared with several traditional protein biomarkers,29 circulating lncRNAs show promising roles as adjunct diagnostic/prognostic epigenetic biomarkers in several disorders, including inflammatory conditions.30–32 In the last years, the circulatory MALAT1 expression profile was screened mainly in adults with sepsis. Pellegrina et al identified that deregulated MALAT1 expression might “contribute to gene expression changes associated with the poorer outcome of elderly patients with sepsis”.33 In the present study, the expression signature of this lncRNA is uncovered for the first time in neonatal sepsis. Our finding could be supported by the vital role MALAT1 plays in regulating the “lipopolysaccharide-induced inflammatory response” via its interaction with nuclear factor (NF)-kappa B.34 It was also implicated in regulating the hyperglycemia-induced inflammatory process.35 Recent findings by Liu et al, in addition, confirm the substantial value of MALAT1 as part of the MALAT1/miR-125 axis in discriminating adult sepsis cases from a healthy population and demonstrate a significant correlation with disease severity, organ injury, and inflammation level.36
Several in vivo and in vitro studies suggested that ANRIL might mediate the inflammatory and immune responses associated with various diseases, including sepsis.10,12,37,38 In line with our findings, recently, Gui et al found overexpression of ANRIL in plasma of non-survivor patients with sepsis than survivors, and the accumulating survival was worse in patients with ANRIL high expression. They proposed that this action could be mediated via enhancing multiple signaling pathways, such as increasing “NLRP3 (NOD-, LRR- and pyrin domain-containing protein 3)” inflammasome by regulating the “miR-122-5p/BRCC3 (BRCA1/BRCA2-Containing Complex Subunit 3) axis”.12
The lncRNA HOTAIR was upregulated and associated with neonatal survival in the present cohort. This finding aligns with the previous experimental studies that revealed the implication of HOTAIR in sepsis progression and outcomes.14,15 Through acting as a microRNA-211 sponge with subsequent interleukin-6 receptor induction, HOTAIR has been found to promote inflammation response, inhibit monocyte proliferation and induce monocyte apoptosis in the mice model of sepsis.13 Furthermore, Shen et al showed that HOTAIR could promote cell apoptosis via the microRNA-22/high mobility group box 1 (HMGB1) pathway in vivo and in vitro.15
Although this study is the first, up to the authors’ knowledge, to explore the implication of the studied lncRNAs in neonatal sepsis, some limitations should be considered. First, the relatively limited sample size, in particular, the healthy control group; Second, the short follow-up duration of hospitalized neonates and absence of testing multiple samples to unravel the stability of the studied lncRNAs in the blood along the progression of the disease. Third, the exploratory nature of the present study lacks explaining the molecular mechanisms by which the studied lncRNAs increase the risk of sepsis or are associated with poor prognosis/survival. Fourth, the specificity of the studied lncRNAs for other neonatal pathological entities is questionable. In this sense, further future studies in large-scale cohorts with long outcome evaluations and molecular experiments are required to unravel the biological significance of these lncRNAs and their potential target genes in neonatal sepsis and the precise underlying molecular mechanisms. Furthermore, replication studies in other neonatal pathological entities, and including pathological controls with multiple sample testing during the disease progress, are recommended to validate the studied lncRNAs specificity in neonatal sepsis.
Conclusion
The present results show upregulation of circulatory lncRNA MALAT1, ANRIL, and HOTAIR in neonatal sepsis compared to healthy neonates. The lncRNAs ANRIL and HOTAIR may have potential prognostic utility as biomarkers for survival in neonatal sepsis.
Data Sharing Statement
All data generated or analyzed during this study are included in this submitted article.
Ethical Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of the Faculty of Medicine, Suez Canal University (approval no. 4463). Informed consent was obtained from all included neonates’ parents or legal guardians before participating in the study.
Acknowledgments
The authors thank all the parents who agreed to let their infants join the study.
Funding
This research received no external funding.
Disclosure
The authors declare no conflicts of interest in relation to this work.
References
1. Sharrow D, Hug L, Liu Y, You DZ. Levels & Trends in Child Mortality Report, 2020. Estimates Developed by the UN Inter-Agency Group for Child Mortality Estimation United Nations Inter-Agency Group for Child Mortality Estimation (UNIGME). World Health Organization; 2020.
2. Li Z, Karlsson O, Kim R, Subramanian SV. Distribution of under-5 deaths in the neonatal, postneonatal, and childhood periods: a multicountry analysis in 64 low-and middle-income countries. Int J Equity Health. 2021;20(1):1–11. doi:10.1186/s12939-021-01449-8
3. Cai S, Thompson DK, Anderson PJ, Yang JY. Short- and long-term neurodevelopmental outcomes of very preterm infants with neonatal sepsis: a systematic review and meta-analysis. Children. 2019;6(12):131. doi:10.3390/children6120131
4. Levy O, Martin S, Eichenwald E, et al. Impaired innate immunity in the newborn: newborn neutrophils are deficient in bactericidal/permeability-increasing protein. Pediatrics. 1999;104(6):1327–1333. doi:10.1542/peds.104.6.1327
5. Levy O. Innate immunity of the human newborn: distinct cytokine responses to LPS and other toll-like receptor agonists. J Endotoxin Res. 2005;11(2):113–116. doi:10.1179/096805105X37376
6. Esposito S, Zampiero A, Pugni L, et al. Genetic polymorphisms and sepsis in premature neonates. PLoS One. 2014;9(7):e101248. doi:10.1371/journal.pone.0101248
7. Ho J, Chan H, Wong SH, et al. The involvement of regulatory non-coding RNAs in sepsis: a systematic review. Crit Care. 2016;20(1):383. doi:10.1186/s13054-016-1555-3
8. Geng F, Liu W, Yu L. Potential role of circulating long noncoding RNA MALAT1 in predicting disease risk, severity, and patients‘ survival in sepsis. J Clin Lab Anal. 2019;33(8):e22968. doi:10.1002/jcla.22968
9. Chen J, He Y, Zhou L, Deng Y, Si L. Long non‑coding RNA MALAT1 serves as an independent predictive biomarker for the diagnosis, severity and prognosis of patients with sepsis. Mol Med Rep. 2020;21(3):1365–1373. doi:10.3892/mmr.2020.10923
10. Hu J, Wang D, Wu H, Yang Z, Yang N, Dong J. Long non-coding RNA ANRIL-mediated inflammation response is involved in protective effect of rhein in uric acid nephropathy rats. Cell Biosci. 2019;9:11. doi:10.1186/s13578-019-0273-3
11. Zhou X, Han X, Wittfeldt A, et al. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-κB pathway. RNA Biol. 2016;13(1):98–108. doi:10.1080/15476286.2015.1122164
12. Gui F, Peng H, Liu Y. Elevated circulating lnc-ANRIL/miR-125a axis level predicts higher risk, more severe disease condition, and worse prognosis of sepsis. J Clin Lab Anal. 2019;33(6):e22917. doi:10.1002/jcla.22917
13. Chen J, Gu X, Zhou L, et al. Long non-coding RNA-HOTAIR promotes the progression of sepsis by acting as a sponge of miR-211 to induce IL-6R expression. Exp Ther Med. 2019;18(5):3959–3967. doi:10.3892/etm.2019.8063
14. Jiang ZJ, Zhang MY, Fan ZW, Sun WL, Tang Y. Influence of lncRNA HOTAIR on acute kidney injury in sepsis rats through regulating miR-34a/Bcl-2 pathway. Eur Rev Med Pharmacol Sci. 2019;23(8):3512–3519. doi:10.26355/eurrev_201904_17717
15. Shen J, Zhang J, Jiang X, Wang H, Pan G. LncRNA HOX transcript antisense RNA accelerated kidney injury induced by urine-derived sepsis through the miR-22/high mobility group box 1 pathway. Life Sci. 2018;210:185–191. doi:10.1016/j.lfs.2018.08.041
16. Wang W, Yang N, Wen R, Liu CF, Zhang TN. Long noncoding RNA: regulatory mechanisms and therapeutic potential in sepsis. Front Cell Infect Microbiol. 2021;11:563126. doi:10.3389/fcimb.2021.563126
17. Martin RJ, Fanaroff AA, Walsh MC. Fanaroff and Martin’s Neonatal-Perinatal Medicine E-Book: Diseases of the Fetus and Infant. Elsevier Health Sciences; 2019.
18. Molloy EJ, Wynn JL, Bliss J, et al. Neonatal sepsis: need for consensus definition, collaboration and core outcomes. Pediatr Res. 2020;88(1):2–4. doi:10.1038/s41390-020-0850-5
19. AbdAllah NB, Toraih EA, Al Ageeli E, et al. MYD88, NFKB1, and IL6 transcripts overexpression are associated with poor outcomes and short survival in neonatal sepsis. Sci Rep. 2021;11(1):13374. doi:10.1038/s41598-021-92912-7
20. Rodgers A, Singh C. Specialist neonatal respiratory care for babies born preterm (NICE guideline 124): a review. Arch Dis Child Educ Pract Ed. 2020;105(6):355–357. doi:10.1136/archdischild-2019-317461
21. Bhandari V. Effective biomarkers for diagnosis of neonatal sepsis. J Pediatric Infect Dis Soc. 2014;3(3):234–245. doi:10.1093/jpids/piu063
22. Toraih EA, Abdelghany AA, Abd El Fadeal NM, Al Ageeli E, Fawzy MS. Deciphering the role of circulating lncRNAs: RNCR2, NEAT2, CDKN2B-AS1, and PVT1 and the possible prediction of anti-VEGF treatment outcomes in diabetic retinopathy patients. Graefes Arch Clin Exp Ophthalmol. 2019;257(9):1897–1913. doi:10.1007/s00417-019-04409-9
23. Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi:10.1373/clinchem.2008.112797
24. Fakhr-Eldeen A, Toraih EA, Fawzy MS. Long non-coding RNAs MALAT1, MIAT and ANRIL gene expression profiles in beta-thalassemia patients: a cross-sectional analysis. Hematology. 2019;24(1):308–317. doi:10.1080/16078454.2019.1570616
25. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-delta delta C (T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
26. Fleischmann C, Reichert F, Cassini A, et al. Global incidence and mortality of neonatal sepsis: a systematic review and meta-analysis. Arch Dis Child. 2021;106(8):745–752. doi:10.1136/archdischild-2020-320217
27. Shepherd R, Cheung AS, Pang K, Saffery R, Novakovic B. Sexual dimorphism in innate immunity: the role of sex hormones and epigenetics. Front Immunol. 2020;11:604000. doi:10.3389/fimmu.2020.604000
28. Everhardt Queen A, Moerdyk-Schauwecker M, McKee LM, Leamy LJ, Huet YM. Differential expression of inflammatory cytokines and stress genes in male and female mice in response to a lipopolysaccharide challenge. PLoS One. 2016;11(4):e0152289. doi:10.1371/journal.pone.0152289
29. Shi T, Gao G, Cao Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers. 2016;2016:9085195. doi:10.1155/2016/9085195
30. Gu Z, Shen HQ, Fu PH, Chen M. Screening of long non-coding RNAs markers in plasma of children with chronic gastritis. Chronic Dis Transl Med. 2020;6(1):62–68. doi:10.1016/j.cdtm.2020.01.001
31. Hashemian SM, Pourhanifeh MH, Fadaei S, Velayati AA, Mirzaei H, Hamblin MR. Non-coding RNAs and exosomes: their role in the pathogenesis of sepsis. Mol Ther Nucleic Acids. 2020;21:51–74. doi:10.1016/j.omtn.2020.05.012
32. Wang C, Liang G, Shen J, et al. Long non-coding RNAs as biomarkers and therapeutic targets in sepsis. Front Immunol. 2021;12:722004. doi:10.3389/fimmu.2021.722004
33. Pellegrina DVDS, Severino P, Barbeiro HV, et al. Insights into the function of long noncoding RNAs in sepsis revealed by gene co-expression network analysis. Noncoding RNA. 2017;3(1):Jan. doi:10.3390/ncrna3010005
34. Zhao G, Su Z, Song D, Mao Y, Mao X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-κB. FEBS Lett. 2016;590(17):2884–2895. doi:10.1002/1873-3468.12315
35. Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med. 2015;19(6):1418–1425. doi:10.1111/jcmm.12576
36. Liu W, Geng F, Yu L. Long non-coding RNA MALAT1/microRNA 125a axis presents excellent value in discriminating sepsis patients and exhibits positive association with general disease severity, organ injury, inflammation level, and mortality in sepsis patients. J Clin Lab Anal. 2020;34(6):e23222. doi:10.1002/jcla.23222
37. Qiao C, Yang L, Wan J, et al. Long noncoding RNA ANRIL contributes to the development of ulcerative colitis by miR-323b-5p/TLR4/MyD88/NF-κB pathway. Biochem Biophys Res Commun. 2019;508(1):217–224. doi:10.1016/j.bbrc.2018.11.100
38. Hu J, Wu H, Wang D, Yang Z, Dong J. LncRNA ANRIL promotes NLRP3 inflammasome activation in uric acid nephropathy through miR-122-5p/BRCC3 axis. Biochimie. 2019;157:102–110. doi:10.1016/j.biochi.2018.10.011
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.