Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 16
Long Non-Coding RNA ZSCAN16-AS1 Promotes the Malignant Progression of Melanoma Through Regulating the miR-503-5p/ARL2 Axis
Authors Zhao Y, Zhang X, Wang J, Li Y, Wu Y, Liu J
Received 25 March 2023
Accepted for publication 16 June 2023
Published 17 July 2023 Volume 2023:16 Pages 1821—1831
DOI https://doi.org/10.2147/CCID.S407323
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Dr Jeffrey Weinberg
Yuting Zhao,1 Xiangzhou Zhang,1 Jie Wang,2 Yong Li,1 Yitong Wu,1 Jisong Liu1
1Department of Plastic Surgery, the Third the People’s Hospital of Bengbu, Bengbu, Anhui, People’s Republic of China; 2Department of Plastic Surgery, Affiliated People’s Hospital of Jiangsu University, Zhenjiang, Jiangsu, People’s Republic of China
Correspondence: Jisong Liu, Department of Plastic Surgery, the Third the People’s Hospital of Bengbu, 38 Shengli Road, Bengbu, Anhui, 233099, People’s Republic of China, Email [email protected]
Background: LncRNA zinc finger and SCAN domain containing 16 antisense RNA 1 (ZSCAN16-AS1), a newly identified lncRNA, has been proven to accelerate hepatocellular carcinoma progression. However, the function and molecular mechanism of ZSCAN16-AS1 in melanoma are still unknown.
Methods: The level of ZSCAN16-AS1 in melanoma tissues was detected and reported in The Cancer Genome Atlas (TCGA) and GEO#GSE15605. CCK-8, Transwell and flow cytometry assays were used to explore the role of ZSCAN16-AS1 in melanoma cells. Luciferase reporter assays and RNA pull-down assays were used to verify the molecular mechanism of ZSCAN16-AS1.
Results: Here, we found that ZSCAN16-AS1 expression was increased in melanoma. We confirmed that ZSCAN16-AS1 promotes the growth and metastasis of melanoma. ZSCAN16-AS1 exerts its pro-tumour role through sponging of miR-503-5p to liberate ADP-ribosylation factor-like protein 2 (ARL2) mRNA transcripts.
Conclusion: These results demonstrated the role and molecular mechanism of ZSCAN16-AS1 in the occurrence and development of melanoma. Therefore, ZSCAN16-AS1 may be used as a specific biomarker in the diagnosis and treatment of melanoma patients.
Keywords: melanoma, growth and metastasis, ZSCAN16-AS1, miR-503-5p, ARL2
Introduction
Cutaneous melanoma is the leading cause of skin cancer-related death, and its global incidence is increasing tremendously each year.1–3 In 2022, the annual incidence of melanoma in China rose to 16.83/10,000, and the annual death toll was 4369.4 At present, comprehensive treatment strategies have been adopted for melanoma, including surgery, chemotherapy, targeted therapy and immunotherapy, but the prognosis of melanoma remains very poor.5,6 Even in some cases of targeted therapy, acquired drug resistance occurs frequently.7 The main reason is the lack of critical biomarkers and therapeutic targets in melanoma. Therefore, further study of the mechanism of melanoma genesis and development is important for the diagnosis and treatment of melanoma patients.
It has been proven that very few RNAs are translated into proteins,8 and this kind of RNA without coding ability is called non-coding RNA (ncRNA).9 Long non-coding RNAs (lncRNAs), with a length of over 200 nucleotides, belong to a subgroup of ncRNAs.10 Many studies have shown that lncRNAs are involved in multiple biological processes of cancer, and some specific lncRNAs have been identified in the progression of melanoma.11,12 LncRNA zinc finger and SCAN domain containing 16 antisense RNA 1 (ZSCAN16-AS1), a brand new lncRNA, is highly expressed in hepatocellular carcinoma and expedites hepatocellular carcinoma progression.13,14 It was reported that ZSCAN16-AS1 is overexpressed in melanoma tissues,15 but the function and molecular mechanism of ZSCAN16-AS1 in melanoma are not very clear.
In this paper, we found that the level of ZSCAN16-AS1 was increased in melanoma. Currently, most research on lncRNAs has demonstrated that most lncRNAs act as competitive endogenous RNAs (ceRNAs) to influence tumour progression.16 Specifically, lncRNAs can promote the expression of specific genes by competitively sponging their target miRNAs.17,18 ADP-ribosylation factor-like protein 2 (ARL2), a member of the ADP-ribosylation factor family,19 has been shown to play an oncogenic role in many human malignant cancers.20,21 miR-503-5p has been proven to play a role as a tumour suppressor gene in many cancers. Here, we demonstrated that miR-503-5p plays a role as a tumour suppressor in melanoma by targeting ARL2. We established the scientific hypothesis that ZSCAN16-AS1 can promote the expression of ARL2 by competitively binding to miR-503-5p in melanoma. We confirmed that ZSCAN16-AS1 expedites growth and metastasis of melanoma through regulating miR-503-5p/ARL2 axis. Therefore, ZSCAN16-AS1 may act as a specific biomarker in the diagnosis and treatment of melanoma patients in the future.
Materials and Methods
Human Tissue Samples
We used The Cancer Genome Atlas (TCGA, 461 melanoma tissues) and Genotype-Tissue Expression (GTEx, 558 normal skin tissues) database to analyse the expression of ZSCAN16-AS1 through GEPIA (http://gepia.cancer-pku.cn/). The other published dataset was GEO#GSE15605 (58 melanoma tissues and 16 normal skin tissues). The 458 melanoma patients’ prognostic data from TCGA were also analysed by using GEPIA (http://gepia.cancer-pku.cn/). All human tissue data are from public databases. The study was approved by the human research ethics committee of the Third the People’s Hospital of Bengbu.
Cell Lines and Culture
We obtained malignant melanoma cell lines (CHL-1, A2058 and A375) from the American Type Culture Collection (ATCC, USA) and human epidermal melanocytes (HEMa-LP) from Invitrogen (USA). Malignant melanoma cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA), and human epidermal melanocytes were grown in medium 254 (Invitrogen, USA). All culture medium was supplemented with 10% foetal bovine serum (Invitrogen, USA). The proper atmosphere for cells growth was 37 °C with 5% carbon dioxide.
Plasmids and Oligonucleotides
The inhibitor and mimic of miR-503-5p and small-interfering-RNA (siRNA) that inhibited the expression of ZSCAN16-AS1 were chemically synthesized by GenePharma (Shanghai, China). The following sequences were used: si-ZSCAN16-AS1 5′-TTGTAAAATTGAAATATTTGAAT-3′ and 5′-GGCATACTTAGTTTTACATTTTT-3′. For the ARL2 plasmid, the amplified ATF2 coding sequence was inserted into the pcDNA3.1 vector (Invitrogen, USA).
Quantitative RT‒PCR
After extracting total RNA from related tissues and cells through TRIzol (Invitrogen, USA), reverse transcription kits (Applied Biosystems, CA) were applied to conduct reverse transcription. In the amplification reaction, an ABI StepOnePlus System (Applied Biosystems, CA) was used, and the reaction conditions were set in advance. The primers were as follows: ZSCAN16-AS1 forwards 5′- GGGCTGCAATAAAACAGCAAA-3′ and ZSCAN16-AS1 reverse 5′- CAATTTCCTATCCCGACCCTCT-3′; ARL2 forwards 5′- GAGCACCGCGGATTCAA-3′ and ARL2 reverse 5′- GCAAAGATGAGGAGGGTTCG-3′. Specific primers for miR-503-5p were obtained from RiboBio (Guangzhou, China). GAPDH was used as a control for measuring ZSCAN16-AS1 and ARL2, and U6 was used as a control for miR-503-5p. To assess the relative expression, we used the 2–ΔΔCt method.
Western Blot
RIPA buffer (KenGEN, China) was used to extract total proteins, and bicinchoninic acid (Invitrogen, USA) was used to quantify the extracted protein. Protein was separated by 10% SDS‒PAGE and transferred to polyvinylidene fluoride membranes (Invitrogen, USA). The membrane was blocked in 5% nonfat milk for 1 h and incubated overnight with a diluted primary antibody against ARL2 (1:1000, Abcam, Cambridgeshire, UK) at 4 °C. Next, the membrane was incubated with goat anti-rabbit secondary antibody (Invitrogen, USA) for 2 hours and exposed to chemiluminescence reagents (Invitrogen, USA). A Gal imaging system was used to collect images.
CCK-8 Assay
To detect the proliferation ability of melanoma cells, cell counting kit-8 (CCK-8, Beyotime, Shanghai, China) was used. Melanoma cells were placed into a 96-well-plate with culture medium, and CCK-8 reagent was added to each well at different times (12 h, 24 h, 48 h, 60 h). We incubated the cells with CCK8 reagent for an additional two hours before measuring the results. A microplate reader (Thermo Scientific, USA) was used to detect the absorbance at 450 nm.
Transwell Assay
We first digested and resuspended transfected melanoma cells. In the upper chamber of a Matrigel-coated Transwell (BD Biosciences, USA), transfected cells were placed in serum-free culture medium. In the lower chamber, we added 10% bovine serum containing culture medium as a lure. After 48 h, invasive melanoma cells were stained with crystal violet. The transwell chamber was photographed upside down.
Flow Cytometry Assay
To measure the apoptosis ratio of melanoma cells, an Annexin V-FITC apoptosis detection kit (BD Biosciences, USA) was used. We first digested and resuspended transfected melanoma cells. We used flow cytometry to detect cell apoptosis according to the instructions. The proportion of apoptosis was assessed through Annexin V-FITC and propidium iodide (PI) double staining. The DNA content was measured to detect the distribution of the cell cycle. Cells were fixed overnight and resuspended in a solution with RNase A and PI. The DNA content was measured by flow cytometry.
Luciferase Reporter Assay
We first predicted the binding sites. The 3’-UTR of ARL2 and ZSCAN16-AS1 fragments were inserted into the reporter plasmid, and the mutant was used as the control. After transfection of oligonucleotides and luciferase reporter plasmids, luciferase activity was detected using the Luciferase Reporter Assay System.
Fluorescence in situ Hybridization (FISH)
For the FISH assay, a Fluorescent In Situ Hybridization Kit was purchased from RiboBio (Guangzhou, China). The experimental procedure was carried out according to a previous study.22 RiboBio (Guangzhou, China) was commissioned to synthesize the ZSCAN16-AS1 probe. The cell nucleus was stained with DAPI, and representative images were obtained by using a confocal microscopy.
RNA Pull-Down Assay
RiboBio (Guangzhou, China) was commissioned to chemically synthesize biotinylated miR-503-5p. Biotinylation mutants and biotinylation NC were used as controls, and the mutated sequence was 3′-GACGUCUUGACAAGGGGCAGGUU-5′. The cells were transfected with these biotinylated oligonucleotides and maintained with M-280 streptavidin magnetic beads (Invitrogen, USA) to obtain the bound RNA,23 and then qRT‒PCR was used to measure the level of ZSCAN16-AS1 in bound RNA.
Statistical Analysis
SPSS13.0 (mean ± SD) was used to evaluate the data, and Student’s t-test (comparison for two groups) and one-way analysis of variance (ANOVA, comparison among multiple groups) was used. The group of data were considered statistically significant when p < 0.05.
Results
ZSCAN16-AS1 is Overexpressed in Melanoma and Has Adverse Effects on the Survival of Melanoma Patients
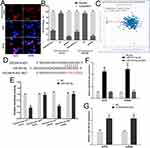
We detected the level of ZSCAN16-AS1 in 461 melanoma tissues and 558 normal skin tissues in the public melanoma datasets of TCGA through the GEPIA website (http://gepia.cancer-pku.cn/index.html). We found that the ZSCAN16-AS1 level was higher in melanoma tissues than in normal skin tissues (Figure 1A). The other public melanoma database of GEO#GSE15605 (including 58 melanoma tissues and 16 normal skin tissues) was used to further measure the expression of ZSCAN16-AS1, and we discovered the same result (Figure 1B). We next confirmed that, compared to human epidermal melanocytes (HEMa-LP), the level of ZSCAN16-AS1 was tremendously increased in malignant melanoma cell lines (CHL-1, A2058 and A375) (Figure 1C). Meanwhile, as shown in Figure 1D, the survival rate of melanoma patients with high ZSCAN16-AS1 levels (expression ratio ≥ median ratio) was poorer by analysing the melanoma prognostic database of TCGA through the GEPIA website.
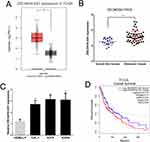 |
Figure 1 ZSCAN16-AS1 is overexpressed in melanoma, and has adverse effects on the survival of melanoma patients. (A) GEPIA (http://gepia.cancer-pku.cn/index.html) was used to detect the expression of ZSCAN16-AS1 in TCGA melanoma dataset. (B) The level of ZSCAN16-AS1 was analyzed in other public melanoma database of GEO#GSE15605 (including 58 melanoma tissues and 16 normal skin tissues). (C) The expression profile of ZSCAN16-AS1 in melanocytes (HEMa-LP) and malignant melanoma cell lines (CHL-1, A2058 and A375). (D) The survival rate of melanoma patients in TCGA was analyzed by using GEPIA (http://gepia.cancer-pku.cn/index.html). *P < 0.05, **P < 0.01, ***P < 0.001. |
ZSCAN16-AS1 Directly Binds to miR-503-5p in Melanoma
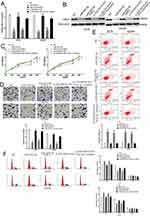
We further studied the molecular mechanism of ZSCAN16-AS1 in melanoma. We used FISH assays and qRT‒PCR of nuclear and cytoplasmic fragments to identify the cellular location of ZSCAN16-AS1, and discovered that ZSCAN16-AS1 was located in both the nucleus and cytoplasm of melanoma cells (Figure 2A and B). lncRNAs have been well proven to function as ceRNAs that can competitively bind to miRNAs in many tumours. LncBase v3.0 (https://diana.e-ce.uth.gr/lncbasev3/interactions) was used to search for specific miRNAs that have binding sites in ZSCAN16-AS1, and 31 miRNAs were found to have potential binding sites in ZSCAN16-AS1 (Supplementary Table 1). Among these miRNAs, miR-503-5p was selected for further study because it is the only miRNA that has a negative correlation with ZSCAN16-AS1 in melanoma based on analysis of the data from TCGA (Figure 2C). We next constructed a ZSCAN16-AS1 luciferase reporter vector that contains the binding sites of miR-503-5p, and mutant vectors were used as the control (Figure 2D). The Activity of luciferase in the wild-type ZSCAN16-AS1 luciferase reporter vector was suppressed by the miR-503-5p mimic, but no significant changes were found in the mutant plasmid (Figure 2E). In addition, an RNA pull-down assay indicated that ZSCAN16-AS1 was pulled down by biotinylated miR-503-5p in melanoma cells (Figure 2F). Endogenetic miR-503-5p expression increased substantially after depletion of ZSCAN16-AS1 in melanoma cells (Figure 2G). All of these results demonstrated that miR-503-5p and ZSCAN16-AS1 have a direct binding relationship in melanoma.
ZSCAN16-AS1 Promotes ARL2 Expression by Sponging miR-503-5p in Melanoma Cells
To identify the target gene of miR-503-5p, bioinformatics software (TarBase, TargetScan, StarBase and miRDB) was used (Figure 3A). ARL2 was found to be a potential target of miR-503-5p. We chose ARL2 for further research because it is the only gene among the 11 potential target genes that has a positive correlation with ZSCAN16-AS1 (Figure 3G). As shown in Figure 3B, the 3’-UTR of ARL2 shares the same binding sites of miR-503-5p with ZSCAN16-AS1. A luciferase reporter assay showed that the luciferase activity of the wild-type ARL2 reporter plasmid was repressed by the miR-503-5p mimic in melanoma cells (Figure 3C). Meanwhile, we also found that the mRNA and protein expression of ARL2 were inhibited by the miR-503-5p mimic (Figure 3D and E). All these results demonstrated that ARL2 is a target gene of miR-503-5p. Furthermore, we also showed that the luciferase activity of the wild-type ARL2 reporter plasmid decreased in the ZSCAN16-AS1 knockdown group, and this effect was reversed by co-transfection with an inhibitor of miR-503-5p (Figure 3F). ARL2 mRNA levels had a positive correlation with ZSCAN16-AS1 in melanoma based on TCGA data (Figure 3G). The protein and mRNA expression of ARL2 declined sharply in the ZSCAN16-AS1 knockdown group, and these suppressed effects could also be abolished by the inhibitor of miR-503-5p (Figure 4A and B). All these results demonstrated that ZSCAN16-AS1 facilitates the expression of ARL2 by sponging miR-503-5p.
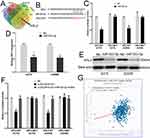 |
Figure 3 ZSCAN16-AS1 promotes ARL2 expression through sponging miR-503-5p in melanoma cells. (A) Bioinformatics software (TarBase, https://dianalab.e-ce.uth.gr/html/diana/web/index.php?r=tarbasev8%2Findex; TargetScan, https://www.targetscan.org/vert_80/; StarBase, https://rnasysu.com/encori/agoClipRNA.php?source=mRNA and miRDB, https://mirdb.org/) was used to identify the target gene of miR-503-5p. (B) The binding sites of miR-503-5p within the 3’-UTR of ARL2. (C) Luciferase activity of melanoma cells transfected with ARL2-WT or ARL2-MUT reporter together with miR-503-5p or NC. (D) The expression of ARL2 mRNA in melanoma cells transfected with miR-503-5p or NC. (E) Western blots identified ARL2 protein expression changes; beta-actin was used as a control. (F) Luciferase assay of melanoma cells transfected with ARL2-WT or ARL2-MUT plasmid together with ZSCAN16-AS1 siRNA or ZSCAN16-AS1 siRNA plus miR-503-5p inhibitor. (G) TCGA melanoma dataset revealed a significant positive correlation between ARL2 mRNA and ZSCAN16-AS1. **P < 0.01. |
ZSCAN16-AS1 Enhances the Growth and Metastasis of Melanoma Cells by Regulating the miR-503-5p/ARL2 Axis
To further explore the biological role of ZSCAN16-AS1 in melanoma cells, ZSCAN16-AS1 siRNA, miR-503-5p mimic, ZSCAN16-AS1 siRNA plus miR-503-5p inhibitor and ARL2 plasmid plus miR-503-5p mimic were transfected into A375 and A2058 cells. The changes in the protein and mRNA expression of ARL2 are shown in Figure 4A and B. The CCK8 assay demonstrated that the growth of melanoma cells was tremendously decreased after downregulating ZSCAN16-AS1 levels or upregulating miR-503-5p levels (Figure 4C). Meanwhile, ZSCAN16-AS1 siRNA and miR-503-5p mimic transfected melanoma cells showed significant inhibition of invasion and an increase in the early apoptosis rate (Figure 4D and E). Flow cytometry showed that the ZSCAN16-AS1 siRNA and miR-503-5p mimic induced cell cycle arrest in G1 phase and decreased the percentage of melanoma cells in S phase (Figure 4F). In addition, the ARL2 plasmid abolished the effect of miR-503-5p (Figure 4C-F), which indicated that miR-503-5p plays a role as a tumour suppressor in melanoma by targeting ARL2. More importantly, the effect of ZSCAN16-AS1 siRNA on the ARL2 level, metastasis, apoptosis, cell cycle and growth of melanoma cells was also reversed by co-transfection with an inhibitor of miR-503-5p (Figure 4C-F). In summary, these results demonstrated that ZSCAN16-AS1 exerts its oncogenic effect in melanoma by regulating the miR-503-5p/ARL2 axis.
Discussion
ncRNAs, a class of RNAs without coding ability, exert their biological role by regulating gene expression at the transcriptional and posttranscriptional levels.24,25 LncRNAs are a class of ncRNAs with a length of more than 200 nucleotides and have been shown to participate in many biological processes, especially in the malignant progression of human cancer.26,27 To date, many specific lncRNAs have been identified that are abnormally expressed and play special roles in many human malignant tumours.28,29 ZSCAN16-AS1, a newly recognized lncRNA, is over-regulated in liver cancer and promotes the malignant properties of hepatocellular carcinoma.13,14 It has been reported that it is also highly expressed in malignant melanoma.15 However, no one has studied the specific function and biological mechanism of ZSCAN16-AS1 in melanoma. Here, we demonstrated that ZSCAN16-AS1 is overexpressed in melanoma by analysing the TCGA and GEO#GSE15605 melanoma databases. High ZSCAN16-AS1 expression has adverse effects on the survival of melanoma patients. We also found that ZSCAN16-AS1 expedites the growth and metastasis of melanoma cells. Next, we further explored the molecular mechanism of ZSCAN16-AS1 in melanoma.
The mechanisms of action of lncRNAs are diverse, including chromatin modifications, transcriptional regulation, RNA processing, mRNA stability and translation.30 To date, it has been demonstrated that lncRNAs can directly interact with proteins to influence their stability and localization, and they also function as ceRNAs in many malignant tumours.31 ceRNA, a kind of RNA that contains miRNA binding sites, reduces the interaction between miRNA and its target genes by competitively binding to miRNA, and regulates the expression of specific oncogenes and tumour suppressors.32 Similarly, ZSCAN16-AS1 has been demonstrated to expedite the malignant progression of hepatocellular carcinoma by modulating the miR-181c-5p/SPAG9/JNK and miR-451a/ATF2 pathways.13,14 Here, we used bioinformatics software and the TCGA database to identify the ceRNA network of ZSCAN16-AS1 in melanoma, and found that ZSCAN16-AS1, miR-503-5p and ARL2 have potential ceRNA correlations. By using luciferase reporter and RNA pull-down assays, we proved that ARL2 is a direct target gene of miR-503-5p and that miR-503-5p and ZSCAN16-AS1 have a direct binding relationship in melanoma. We also demonstrated that ZSCAN16-AS1 facilitates the expression of ARL2 through decaying miR-503-5p in melanoma.
ARL2, a member of the ADP-ribosylation factor family, is a small G-protein.19 Small G-proteins participate in multiple cellular signal transduction pathways and functions, including cytoskeleton construction, cell differentiation and exosome transport.33 ARL2 binds to tubulin cofactor D (TBCD) in the form of ARL2-GDP in the cytoplasm to regulate microtubule dynamics.34 The complex formed by ARL2 GTP and ARL2 binding factor (BART) enters mitochondria and maintains mitochondrial morphology, movement, ATP levels, and so on.35,36 In addition, ARL2 can enhance the interaction between BART and STAT3, promoting STAT3 nuclear translocation.37 ARL2 in cancer progression has been documented and has been shown to play an oncogenic role in many human malignant cancers, such as cervical cancer hepatocellular carcinoma, pancreatic cancer and breast cancer.20,21,38,39 For instance, ARL2 knockdown contributed to cell apoptosis and impeded the proliferation of osteosarcoma cells.40 Overexpression of ARL2 almost abrogated PVT1 deficiency-mediated anti-proliferation, pro-apoptosis, and anti-metastasis effects on cervical cancer cells.41 miR-503-5p has been proven to play a role as a tumour suppressor gene in many cancers, including colon cancer and osteosarcoma.42,43 In this paper, we demonstrated that miR-503-5p also plays an anti-cancer role in melanoma through targeted inhibition of ARL2 expression. Moreover, we showed that co-transfection of an inhibitor of miR-503-5p abolished the effect of ZSCAN16-AS1 siRNA on the ARL2 level, metastasis, apoptosis, cell cycle and growth of melanoma cells. In summary, ZSCAN16-AS1 is crucial to the malignant progression of melanoma. This study indicated that ZSCAN16-AS1 exerts its oncogenic effect in melanoma by regulating the miR-503-5p/ARL2 axis. ZSCAN16-AS1 can be a novel potential target for melanoma treatment in the future.
Conclusion
In conclusion, we demonstrated that ZSCAN16-AS1 prevents the binding of miR-503-5p and ARL2 by decoying miR-503-5p, resulting in increased ARL2 expression. ZSCAN16-AS1 acts as an oncogene in melanoma by regulating the miR-503-5p/ARL2 axis. Understanding the internal molecular mechanism of the ZSCAN16-AS1/miR-503-5p/ARL2 network in melanoma will be beneficial to identify new and meaningful therapeutic targets for patients with melanoma. It has been reported that the combination of radiotherapy and immunotherapy seems to be a safe therapeutic option and shows better results in terms of survival outcomes in melanoma.44 Therefore, it is meaningful for researchers to explore the role of the ZSCAN16-AS1/miR-503-5p/ARL2 network in terms of radiotherapy and immunotherapy for melanoma patients in the future.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Ahmed B, Qadir MI, Ghafoor S. Malignant melanoma: skin cancer-diagnosis, prevention, and treatment. Crit Rev Eukaryot Gene Expr. 2020;30(4):291–297.
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30.
3. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
4. Xia C, Dong X, Li H, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J. 2022;135(5):584–590.
5. Kozovska Z, Gabrisova V, Kucerova L. Malignant melanoma: diagnosis, treatment and cancer stem cells. Neoplasma. 2016;63(4):510–517.
6. Paluncic J, Kovacevic Z, Jansson PJ, et al. Roads to melanoma: key pathways and emerging players in melanoma progression and oncogenic signaling. Biochim Biophys Acta. 2016;1863(4):770–784.
7. Lu H, Liu S, Zhang G, et al. PAK signalling drives acquired drug resistance to MAPK inhibitors in BRAF-mutant melanomas. Nature. 2017;550(7674):133–136.
8. Elgar G, Vavouri T. Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet. 2008;24(7):344–352.
9. Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7(8):8601–8612.
10. Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21(5):542–551.
11. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719.
12. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346.
13. Liu J, Liu R, Liu Y, et al. ZSCAN16-AS1 expedites hepatocellular carcinoma progression via modulating the miR-181c-5p/SPAG9 axis to activate the JNK pathway. Cell Cycle. 2021;20(12):1134–1146.
14. Lv C, Wan Q, Shen C, Wu H, Zhou B, Wang W. Long non‑coding RNA ZSCAN16‑AS1 promotes the malignant properties of hepatocellular carcinoma by decoying microRNA‑451a and consequently increasing ATF2 expression. Mol Med Rep. 2021;24(5):67.
15. Ding Y, Li M, Tayier T, Zhang M, Chen L, Feng S. Bioinformatics analysis of lncRNA‑associated ceRNA network in melanoma. J Cancer. 2021;12(10):2921–2932.
16. Cheng JT, Wang L, Wang H, et al. Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition. Cells. 2019;8(10):35.
17. Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;87:15–24.
18. Lingadahalli S, Jadhao S, Sung YY, et al. Novel lncRNA LINC00844 Regulates Prostate Cancer Cell Migration and Invasion through AR Signaling. Mol Cancer Res. 2018;16(12):1865–1878.
19. Wang Y, Guan G, Cheng W, et al. ARL2 overexpression inhibits glioma proliferation and tumorigenicity via down-regulating AXL. BMC Cancer. 2018;18(1):599.
20. Chen L, Zhang X, Wang S, Lin X, Xu L. Circ_0084927 Facilitates Cervical Cancer Development via Sponging miR-142-3p and Upregulating ARL2. Cancer Manag Res. 2020;12:9271–9283.
21. Beghin A, Belin S, Hage-Sleiman R, et al. ADP ribosylation factor like 2 (Arl2) regulates breast tumor aggressivity in immunodeficient mice. PLoS One. 2009;4(10):e7478.
22. Liu H, Dai C, Wu Q, Liu H, Li F. Expression profiling of long noncoding RNA identifies lnc-MMP3-1 as a prognostic biomarker in external auditory canal squamous cell carcinoma. Cancer Med. 2017;6(11):2541–2551.
23. Subramanian M, Li XL, Hara T, Lal A. A biochemical approach to identify direct microRNA targets. Methods Mol Biol. 2015;1206:29–37.
24. Zhang X, Wang W, Zhu W, et al. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int J Mol Sci. 2019;20(22):85.
25. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29(4):452–463.
26. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914.
27. Xing YH, Bai Z, Liu CX, Hu SB, Ruan M, Chen LL. Research progress of long noncoding RNA in China. IUBMB Life. 2016;68(11):887–893.
28. Liu L, Shi Y, Shi J, et al. The long non-coding RNA SNHG1 promotes glioma progression by competitively binding to miR-194 to regulate PHLDA1 expression. Cell Death Dis. 2019;10(6):463.
29. Pandey GK, Kanduri C. Long Non-Coding RNAs: tools for Understanding and Targeting Cancer Pathways. Cancers. 2022;14(19):4760.
30. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220(2).:56
31. Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369.
32. Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54(5):766–776.
33. Matozaki T, Nakanishi H, Takai Y. Small G-protein networks: their crosstalk and signal cascades. Cell Signal. 2000;12(8):515–524.
34. Bhamidipati A, Lewis SA, Cowan NJ. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J Cell Biol. 2000;149(5):1087–1096.
35. Sharer JD, Shern JF, Van Valkenburgh H, Wallace DC, Kahn RA. ARL2 and BART enter mitochondria and bind the adenine nucleotide transporter. Mol Biol Cell. 2002;13(1):71–83.
36. Newman LE, Zhou CJ, Mudigonda S, et al. The ARL2 GTPase is required for mitochondrial morphology, motility, and maintenance of ATP levels. PLoS One. 2014;9(6):e99270.
37. Muromoto R, Sekine Y, Imoto S, et al. BART is essential for nuclear retention of STAT3. Int Immunol. 2008;20(3):395–403.
38. Hass HG, Vogel U, Scheurlen M, Jobst J. Gene-expression Analysis Identifies Specific Patterns of Dysregulated Molecular Pathways and Genetic Subgroups of Human Hepatocellular Carcinoma. Anticancer Res. 2016;36(10):5087–5095.
39. Taniuchi K, Iwasaki S, Saibara T. BART inhibits pancreatic cancer cell invasion by inhibiting ARL2-mediated RhoA inactivation. Int J Oncol. 2011;39(5):1243–1252.
40. Sun Z, Li A, Yu Z, Li X, Guo X, Chen R. MicroRNA-497-5p Suppresses Tumor Cell Growth of Osteosarcoma by Targeting ADP Ribosylation Factor-Like Protein 2. Cancer Biother Radiopharm. 2017;32(10):371–378.
41. Liu W, Yao D, Huang B. LncRNA PVT1 promotes cervical cancer progression by sponging miR-503 to upregulate ARL2 expression. Open Life Sci. 2021;16(1):1–13.
42. Wei L, Sun C, Zhang Y, Han N, Sun S. miR-503-5p inhibits colon cancer tumorigenesis, angiogenesis, and lymphangiogenesis by directly downregulating VEGF-A. Gene Ther. 2022;29(1–2):28–40.
43. Li J, Zhang F, Li H, et al. Circ_0010220-mediated miR-503-5p/CDCA4 axis contributes to osteosarcoma progression tumorigenesis. Gene. 2020;763:145068.
44. Tagliaferri L, Lancellotta V, Fionda B, et al. Immunotherapy and radiotherapy in melanoma: a multidisciplinary comprehensive review. Hum Vaccin Immunother. 2022;18(3):1903827.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.