Back to Journals » OncoTargets and Therapy » Volume 12
Long non-coding RNA XIST promotes malignant behavior of epithelial ovarian cancer
Authors Zuo K, Zhao Y, Zheng Y, Chen D, Liu X, Du S, Liu Q
Received 6 February 2019
Accepted for publication 16 July 2019
Published 5 September 2019 Volume 2019:12 Pages 7261—7267
DOI https://doi.org/10.2147/OTT.S204369
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Kun Zuo1,*, Youhong Zhao1,*, Yukun Zheng2, De Chen1, Xiaoli Liu1, Song Du1, Qing Liu1
1Gansu Provincial Maternity and Child-care Hospital, Lanzhou 730050, People’s Republic of China; 2Department of Obstetrics and Gynecology, The First Hospital of Lanzhou University, Lanzhou 730000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qing Liu
Gansu Provincial Maternity and Child-care Hospital, No. 143 Qilihe North Street, Lanzhou 730050, People’s Republic of China
Tel +86 0 931 518 8888
Email [email protected]
Purpose: This study aims to investigate the functional role of long non-coding RNA XIST in epithelial ovarian cancer (EOC).
Methods: Detection of XIST expression levels in EOC tissues and cell lines was done using qRT-PCR. The relationship between XIST expression and clinicopathological features of EOC patients was compared and analyzed. The cumulative survival rates were calculated using Kaplan-Meier. A Cox hazard model was used to identify risk factors for survival. Lastly, the effects of XIST on EOC cell were assessed in vitro.
Results: XIST was up-regulated in EOC tissues and cell lines. The expression of XIST was closely related to the tumor grade, distant metastasis, and FIGO stage in the EOC patients. The Cox regression analysis showed that high XIST expression was an independent predictor of prognosis in patients with EOC. In in vitro experiments, reducing XIST expression significantly suppressed cell proliferation, migration and invasion in EOC cells.
Conclusion: XIST highly expressed in the EOC and plays a role in tumor promotion, which may be a potential target for the treatment of EOC.
Keywords: XIST, EOC, prognosis, proliferation, migration, invasion
Introduction
Ovarian cancer is one of the most common malignant tumors in females, which greatly affects the health of women around the world. According to the latest statistics, US is witnessing a total of 22,240 new ovarian cancer cases and about 14,070 ovarian cancer-related deaths annually.1 Based on the histological characteristics, ovarian cancer can be classified into epithelial, stromal secretory, and germ cell types. Among these, epithelial ovarian cancer (EOC) is the most common type of ovarian cancer and accounts for around 90% of all ovarian cancer cases.2 The comprehensive antitumor pattern has developed rapidly, however the survival time of EOC patients has not improved. Therefore, the molecular mechanisms involved in the development of EOC and effective therapeutic targets are current research hotspots.
In recent years, long non-coding RNAs (lncRNAs) have been emerged as research hotspots in the field of tumor biology. lncRNAs are transcripts >200 nucleotides in length, which do not have protein-coding functions.3 lncRNAs are involved in multiple important regulatory processes, such as silencing of chromosome X, genome blotting and chromatin modification, transcription activation, transcription interference, and intranuclear transport.4–6 lncRNAs have characteristic expression patterns in tumors and other diseases. Among them, the lncRNA X-inactive specific transcript (XIST) plays an important role in X chromosome inactivation, which is closely associated with tumor formation. XIST is involved in numerous aspects of cancer genesis, including tumor initiation, invasion, metastasis, cell apoptosis, cell cycle, stemness, autophagy, and drug resistance.7 Moreover, XIST is also known to participate in the progression of several malignant solid tumors, such as gastric cancer, colorectal cancer, retinoblastoma, lung cancer, and liver cancer.8–12 However, research related to XIST and EOC has been rarely carried out. In this study, we aimed to elucidate the role of XIST in EOC using EOC tissues and cell lines.
Materials and methods
Tissue collection
Ninety-eight pairs of surgically resected EOC tissues and the adjacent tissues were collected at Gansu Provincial Maternity and Child Care Hospital, China from January 2010 to December 2012. All the 98 EOC patients were pathologically verified to have the tumor and complete clinicopathological data were obtained from them. All patients received the FIGO stage. The tissue specimens were frozen in liquid nitrogen and stored at –80 °C until further use. All the patients were naive to chemotherapy, radiotherapy, and immunotherapy before surgery and provided written informed consent for using their tissues in this study. This study was performed in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Gansu Provincial Maternity and Child Care Hospital, China.
Cell culture and transfection
Human EOC cell lines (OVCAR3, OV90, A2780, and SKOV3) and human ovarian surface epithelial cell line (HOSE) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). All the cell lines were cultured in RPMI-1640 medium (Gibco-BRL, Gaithersburg, MD, USA)containing 10% fetal bovine serum (FBS, HyClone, Logan, Utah, USA) and 100 Units/mL penicillin-streptomycin (Invitrogen, Carlsbad, CA, USA) at 37 °C in a humidified environment. The two siRNA sequences targeting XIST were as follows: siRNA1: 5ʹ-GCTTCTAACTAGCCTGAAT-3ʹ, and siRNA2: 5ʹ-GCATGCATCTTGGACATTT-3ʹ. Subsequently, cells were cultured in a 6-well plate overnight and transfected with 50 nM siRNA1 or siRNA2 using Lipofectamine RNAiMAX Reagent (Invitrogen, Breda, The Netherlands) in accordance with the manufacturer’s instructions. Non-transfected cells were used as the negative control group (NC group).
qRT-PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA was reverse transcribed into cDNA using Reverse Transcription Kit (Takara, Dalian, China’s Republic of China) and qRT-PCR was performed using SYBR Premix Ex Taq II (Takara, Dalian, China). The reaction conditions were as follows: enzyme activation at 95 °C for 5 min, denaturation at 95 °C for 20 s, annealing at 60 °C for 30 s, and extension at 72 °C for 20 s for 45 cycles. The expression levels were normalized with GAPDH, and the relative expression of lncRNA was calculated according to the 2− ΔΔCT method. The primer sequences of XIST used were as follows: sense, 5ʹ-CTTGGATGGGTTGCCAGCTA-3ʹ; antisense, 3ʹ-TCATGCCCCATCTCCACCTA-5ʹ.
CCK-8
Cell proliferation was determined using Cell Counting Kit-8 (CCK8; Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Briefly, cells were seeded in a 96-well plate at a density of 1×104 cells/well overnight. Subsequently, the corresponding siRNAs were transfected into the cells and cultured in the normal medium. After 1, 2, 3, 4 and 5 days of transfection, 10 μL of CCK-8 reagent was added into each well, followed by incubation for 1 h. Finally, the absorbance of each well was recorded at a wavelength of 450 nm using a microplate reader (Bio-Rad, Hercules, CA, USA).
Transwell assay
Cell migration and invasion were detected using a Transwell assay. For the cell invasion assay, Transwell chambers (8.0 μm, Millipore, Temecula, MA, USA) were coated with 50 mg/L of Matrigel diluent. The cells were cultured in serum-free medium for 24 h after transfection for 48 h. The Transwell chambers were then placed in a 24-well plate containing 10% FBS. In the next step, 100 μL of cell suspension was added into the chamber containing serum-free medium. After 48 h of incubation, cells were washed with PBS, fixed in cold ethanol, and stained with crystal violet solution for 30 min. The number of cells migrating to the lower membrane was counted using an inverted microscope (Olympus Corporation, Tokyo, Japan), and the fields used for counting of migrating or invading cells were selected randomly.
Wound healing assay
Cell migration capacity was detected by wound healing assay. Briefly, 5×105 cells were seeded in a 12-well plate, and after reaching 90% confluency, cells at the bottom surface of wells were scraped using a 10-μL sterile pipette. Subsequently, cells were washed thrice with PBS and incubated for 24 h at 37 °C with 5% CO2. The cell migration rate was analyzed by comparing the width of initial scratch with that after 24 h of wounding under a microscope.
Statistical analysis
The data are expressed as mean ± SD. Differences in the mean between two samples were evaluated by Student’s t-test, and the differences in proportion were evaluated using Fisher’s Exact test or Chi-square test. In this study, progression-free survival (PFS) and overall survival (OS) were used as outcome measures for EOC patients. The cumulative PFS and OS rates were calculated using the Kaplan-Meier method, and the differences were compared using a log-rank test. A Cox proportional hazard regression model was used for the univariate and multivariate analyses. SPSS version 17.0 (SPSS Inc., Chicago, IL, USA) was used for all the analyses. A p-value of less than 0.05 was considered statistically significant.
Results
XIST is overexpressed in EOC
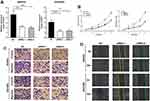
To assess the role of XIST in EOC, we examined the expression levels of XIST in EOC and the adjacent tissues. As the results showed that XIST expression in the EOC tissues was significantly higher than that in the adjacent tissues (Figure 1A). To further explore the role of XIST in EOC, the expression level of XIST was examined in EOC and HOSE cell lines. Expression of XIST in all four EOC cell lines was upregulated (Figure 1B) compared to that in the normal ovarian epithelial HOSE cells.
Further, to analyze the relationship between expression of XIST and the clinicopathological features of EOC, 98 cases of EOC patients were divided into a low (n=46) and high (n=52) expression group according to the median value of XIST expression. As shown in Table 1, the expression of XIST was closely related to the tumor grade, distant metastasis, and FIGO stage in the EOC patients.
 |
Table 1 Correlations between XIST expression and clinicopathological parameters in EOC patients |
XIST expression correlated with poor prognosis of EOC
To investigate the relationship between XIST expression and prognosis of patients with EOC, we performed a long-term follow-up for all the EOC patients. At the end of the follow-up period, survival curves for 98 EOC patients were plotted. As shown in Figure 1C and D, EOC patients with high XIST expression had shorter PFS or OS. We analyzed various variables affecting OS of the EOC patients using the Cox proportional hazard regression model. Univariate analysis showed that the prognosis of EOC patients was closely related to the expression level of XIST. These data suggested that advanced FIGO stage, distant metastasis, and high XIST expression indicate a worse prognosis in the EOC patients. Multivariate analysis showed that high XIST expression is an independent predictor of prognosis in patients with EOC. These results are summarized in Table 2.
 |
Table 2 Univariate and multivariate Cox regression analyses of prognostic factors in EOC patients |
XIST promoted the malignant behavior of EOC cells
It is worth mentioning that among the four EOC cell lines used in this study, SKOV3 and OVCAR3 cells showed the higher XIST expression (Figure 1B). Therefore, we used SKOV3 and OVCAR3 cells for subsequent cellular experiments. We knocked down XIST in the SKOV3 and OVCAR3 cells by transfecting them with XIST siRNA and verified the inhibition efficiency by qRT-PCR (Figure 2A). As shown by the CCK-8 assay, XIST siRNA-transfected SKOV3 or OVCAR3 cells showed reduced cell growth compared to the control non-transfected cells (Figure 2B). In the Transwell assay, invasion and migration ability of cells transfected with XIST siRNA were significantly lower than those in the control cells (Figure 2C). Similar to the results of the above experiments, the wound healing ability of cells transfected with XIST siRNA was also reduced (Figure 2D). Taken together, the above results clearly show that downregulation of XIST inhibits the proliferation, invasion, and migration of SKOV3 or OVCAR3 cells.
Discussion
EOC has the highest malignant grade and shows strong invasion among all the pathological subtypes of ovarian cancer. Such pathological features correspond to rapid disease progression and recurrence after treatment. About 75% of EOC patients have distant metastasis at the time of diagnosis.13 Great progress has been achieved in the surgery-based comprehensive antitumor pattern, and the prognosis for EOC patients has markedly improved, although the 5-year survival rate of advanced EOC patients is only 30%.14 Therefore, it is crucial to investigate the molecular regulatory mechanisms related to the malignant behaviors of EOC and search for new therapeutic targets. lncRNAs are regarded as new regulators of tumor genesis and cancer progression. A variety of lncRNAs have been identified that play an important role in EOC; eg, LINC00460 can promote EOC progression by regulating microRNA-338-3p, whereas the lncRNA HULC can suppress ATG7 and promote EOC by regulating ITGB1.15,16 An Italian multicenter retrospective study suggested that lncRNAs such as SERTAD2-3, SOX4-1, HRCT1-1, and PVT1 could serve as markers to predict the recurrence and poor prognosis of stage I EOC patients.17 Nonetheless, role of XIST in EOC has not been reported so far. Wang et al demonstrated by in vitro experiments that upregulation of the lncRNA XIST can suppress the development of EOC.18 However, for their study, they used only the EOC cell line, and not the normal ovarian epithelial cell line. Our study is the first to report XIST expression in both EOC tissues and cell lines. XIST expression in EOC tissues was markedly upregulated compared to that in the paracarcinoma tissues, and similar results were obtained in the EOC cell lines.
Wang et al demonstrated that XIST was highly expressed in non-small cell lung cancer tissues and cell lines and could promote tumor cell proliferation and invasion by suppressing microRNA-186.19 Besides, XIST also functions as an oncogene in retinoblastoma. Cheng showed that XIST expression was upregulated in both retinoblastoma tissues and cell lines.20 In cell-based experiments, effective knockdown of XIST could inhibit tumor cell proliferation, migration, and invasion while promoting cell apoptosis and caspase-3 activity. They further demonstrated that XIST serves as a ceRNA to competitively absorb miR-101 and promote epithelial-mesenchymal transition. The results of a meta-analysis suggested that XIST expression negatively correlated with the overall survival of patients with malignant solid tumors, and XIST upregulation positively correlated with advanced clinical TNM stage, lymph node metastasis, and distant metastasis.21 Moreover, XIST was also associated with tumor size. Our preliminary results demonstrate that XIST expression is upregulated in EOC, and might promote cancer development during EOC progression. Therefore, we further analyzed the relationship between XIST and the prognosis in EOC patients. The EOC patients showing high XIST expression had poorer prognosis, and XIST was a poor prognostic factor in these patients. In in vitro experiments, we suppressed XIST expression in the EOC cell lines using XIST siRNAs and performed tumor cell functional analysis. Results of the cell experiments corroborated the clinicopathological features and revealed that XIST knockdown could inhibit tumor cell proliferation, migration, and invasion.
Based on the tissue and cell experiments, we conclude that XIST is highly expressed in EOC and functions as an oncogene. However, the study has certain limitations such as lack of animal experiments and incomplete elucidation of the detailed molecular mechanisms. In the future, we plan to perform experiments that will focus on illustrating the related molecular mechanisms to provide new directions for the treatment of EOC.
Ethics approval
This study was conducted after obtaining local ethical committee approval from Gansu Provincial Maternity and Child Care Hospital. All patients signed informed consent, and the study was conducted in accordance with the Declaration of Helsinki.
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30.
2. Morgan RJ
3. Perkel JM. Visiting “noncodarnia”. Biotechniques. 2013;54(6):301, 303–4.
4. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407.
5. Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455.
6. Chen CK, Blanco M, Jackson C, et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354(6311):468–472.
7. Yang Z, Jiang X, Jiang X, Zhao H. X-inactive-specific transcript: a long noncoding RNA with complex roles in human cancers. Gene. 2018;679:28–35.
8. Gao S, Zhao ZY, Wu R, Zhang Y, Zhang ZY. Prognostic value of long noncoding RNAs in gastric cancer: a meta-analysis. Onco Targets Ther. 2018;11:4877–4891.
9. Zhu J, Zhang R, Yang D, et al. Knockdown of long non-coding RNA XIST inhibited doxorubicin resistance in colorectal cancer by upregulation of miR-124 and downregulation of SGK1. Cell Physiol Biochem. 2018;51(1):113–128.
10. Hu C, Liu S, Han M, Wang Y, Xu C. Knockdown of lncRNA XIST inhibits retinoblastoma progression by modulating the miR-124/STAT3 axis. Biomed Pharmacother. 2018;107:547–554.
11. Li C, Wan L, Liu Z, et al. Long non-coding RNA XIST promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018;418:185–195.
12. Chang S, Chen B, Wang X, Wu K, Sun Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017;17(1):248.
13. Liu S, Liu Y, Lu Q, Zhou X, Chen L, Liang W. The lncRNA TUG1 promotes epithelial ovarian cancer cell proliferation and invasion via the WNT/β-catenin pathway. Onco Targets Ther. 2018;11:6845–6851.
14. Jin Y, Feng SJ, Qiu S, Shao N, Zheng JH. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2017;21:3176–3184.
15. Liu X, Wen J, Wang H, Wang Y. Long non-coding RNA LINC00460 promotes epithelial ovarian cancer progression by regulating microRNA-338-3p. Biomed Pharmacother. 2018;108:1022–1028.
16. Chen S, Wu DD, Sang XB, et al. The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis. 2017;8(10):e3118.
17. Martini P, Paracchini L, Caratti G, et al. lncRNAs as novel indicators of patients’ prognosis in stage I epithelial ovarian cancer: a retrospective and multicentric study. Clin Cancer Res. 2017;23(9):2356–2366.
18. Wang C, Qi S, Xie C, Li C, Wang P, Liu D. Upregulation of long non-coding RNA XIST has anticancer effects on epithelial ovarian cancer cells through inverse downregulation of hsa-miR-214-3p. J Gynecol Oncol. 2018;29(6):e99.
19. Wang H, Shen Q, Zhang X, et al. The long non-coding RNA XIST controls non-small cell lung cancer proliferation and invasion by modulating miR-186-5p. Cell Physiol Biochem. 2017;41(6):2221–2229.
20. Cheng Y, Chang Q, Zheng B, Xu J, Li H, Wang R. LncRNA XIST promotes the epithelial to mesenchymal transition of retinoblastoma via sponging miR-101. Eur J Pharmacol. 2019;843:210–216.
21. Zhou Q, Hu W, Zhu W, et al. Long non coding RNA XIST as a prognostic cancer marker - A meta-analysis. Clin Chim Acta. 2018;482:1–7.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.