Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA SPRY4-IT1 Reverses Cisplatin Resistance by Downregulating MPZL-1 via Suppressing EMT in NSCLC
Authors Ye Y , Gu J, Liu P , Wang H, Jiang L, Lei T, Yu S, Han G, Wang Z
Received 2 October 2019
Accepted for publication 8 March 2020
Published 2 April 2020 Volume 2020:13 Pages 2783—2793
DOI https://doi.org/10.2147/OTT.S232769
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Yunyao Ye,1,2,* Jingyao Gu,1,* Pei Liu,1,3,* He Wang,1,4 Lihua Jiang,1 Tianyao Lei,1 Shanxun Yu,2 Gaohua Han,2 Zhaoxia Wang1
1Department of Oncology, The Second Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu, People’s Republic of China; 2Department of Oncology, Taizhou People’s Hospital, Taizhou, Jiangsu, People’s Republic of China; 3Department of Digestive Oncology, The Fourth Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu, People’s Republic of China; 4Department of Oncology, Sir Run Run Hospital, Nanjing Medical University, Nanjing, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhaoxia Wang
Department of Oncology, The Second Affiliated Hospital of Nanjing Medical University, Nanjing 210011, People’s Republic of China
Tel +86-25-58509810
Fax +86-25-58509994
Email [email protected]
Purpose: Long non-coding RNA (lncRNA) SPRY4 intronic transcript 1 (SPRY4-IT1) is reported to play important roles in the occurrence and development of many tumors. However, the possible role of SPRY4-IT1 in cisplatin (DDP) resistance of non-small-cell lung cancer (NSCLC) remains unclear. The aim of this study is to investigate the functions and molecular mechanisms underlying SPRY4-IT1 of cisplatin resistance in NSCLC.
Methods: Expression of SPRY4-IT1 was analyzed in A549 and cisplatin-resistant A549/DDP cell lines by quantitative real-time polymerase chain reaction (RT-qPCR). Overexpression techniques were applied to investigate the biological functions of SPRY4-IT1 in cisplatin-resistant A549/DDP cells. The effects of SPRY4-IT1 on proliferation and apoptosis were evaluated using cell counting kit-8 (CCK8) assays, colony formation assay and flow-cytometric analysis. The expressions of epithelial–mesenchymal transition (EMT)-associated proteins, including E-cadherin and Vimentin, were detected by Western blot. Microarray analysis was performed to identify the putative targets of SPRY4-IT1, which were further verified by Western blotting and RT-qPCR. A549/DDP cells transfected with pCDNA-SPRY4-IT1 were injected into nude mice in order to verify the effect of SPRY4-IT1 on cisplatin resistance in vivo.
Results: The present study demonstrated that SPRY4-IT1 expression was decreased in A549/DDP cells compared with parental A549 cells. Upregulation of SPRY4-IT1 suppressed cell proliferation and caused apoptosis of A549/DDP cells both in vitro and in vivo. MPZL-1 was negatively regulated by SPRY4-IT1. Furthermore, upregulation of SPRY4-IT1 and downregulation of MPZL-1 could suppress epithelial–mesenchymal transition (EMT), which was characterized by increased E-cadherin expression and decreased Vimentin expression.
Conclusion: Upregulation of SPRY4-IT1 reversed the cisplatin-resistant phenotype of NSCLC partially by downregulating MPZL-1 via inhibiting EMT process.
Keywords: long non-coding RNA, SPRY4-IT1, cisplatin resistance, MPZL-1, EMT
Introduction
Lung cancer is one of the most common types of cancer, and the leading cause of cancer-associated mortality in both men and women worldwide.1 Non-small-cell lung cancer (NSCLC) accounts for 80–85% of all lung cancer cases.2 Despite considerable progress in treatment of the disease, platinum-based combination chemotherapy, especially cisplatin,3 still remains the mainstay of NSCLC treatment, for neoadjuvant therapy, adjuvant therapy and palliative therapy. However, cisplatin resistance severely impedes NSCLC treatment and becomes a major cause of the therapy failure. Therefore, understanding the molecular mechanisms underlying chemoresistance is crucial for the effective therapeutic strategy and improvement of survival rate.
Long noncoding RNAs (lncRNAs) are non-protein-coding RNAs, with more than 200 bp in length.4 lncRNAs function as key regulators of several biological processes, such as cell proliferation, apoptosis, migration and invasion by epigenetic, transcriptional and post-transcriptional levels.5 The dysregulated expressions of lncRNAs, acting as oncogenes or antioncogenes, have been demonstrated to be related with the occurrence and progression of a variety of cancers. An accumulating evidence indicates that lncRNAs are also implicated in the development of tumor chemoresistance.6 For instance, Li et al found that highly expressed lncRNA SNHG1 involves in the hepatocellular carcinoma cells sorafenib resistance via activating the Akt pathway (PMID: 31053148). Moreover, LINC00665 contributes to the NSCLC cells acquired resistance to gefitinib by interacting with EZH2 and activating the PI3K/AKT pathway (PMID: 30889481).
LncRNA SPRY4 intronic transcript 1 (SPRY4-IT1) is derived from an intron within SPRY4,7 which plays an important role in melanoma cell growth and invasion. Aberrant expression of SPRY4-IT1 has also been found in many other cancers, such as prostate cancer, breast cancer and esophageal cancer.8,9 Our previous study showed that EZH2-mediated epigenetic suppression of SPRY4-IT1 is correlated with a poor prognosis of NSCLC, and promotes cell proliferation and metastasis by affecting the epithelial–mesenchymal transition (EMT).10 However, whether SPRY4-IT1 influences the development of chemoresistance in NSCLC remains unclear. In this study, we investigated the effects of lncRNA SPRY4-IT1 expression on the chemosensitivity of NSCLC cells to cisplatin both in vitro and in vivo. Furthermore, we clarified for the first time that overexpression SPRY4-IT1 could reverse cisplatin resistance by inhibiting MPZL-1 via inhibiting EMT in NSCLC.
Materials and Methods
Cell Lines and Cell Culture
The cisplatin-resistant human lung cancer cell line (A549/DDP) and its parental cell line (A549) were obtained from the Cancer Institute, Chinese Academy of Sciences. All cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin under 5% CO2 at 37°C. The cisplatin-resistant cell line A549/DDP was exposed to the medium containing 1.0 μg/mL cisplatin in order to maintain cisplatin resistance.
RNA Extraction and RT-qPCR
Total RNA was extracted from cells or transfected cells by using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Total RNA (500 ng) was reverse transcribed by using PrimeScript RT reagent Kit (Takara, China). The RNA expression was determined by SYBR Premix Ex Taq (TaKaRa, Dalian, China) and normalized with the expressions of GAPDH. SPRY4-IT1 expression was determined by qRT-PCR using the following primer sequences: sense, 5ʹ-AGCCACATAAATTCAGCAGA-3ʹ; and reverse, 5ʹ-CGATGTAGGATTCCTTTCA-3ʹ. The primer for MPZL-1 was designed as: sense, 5ʹ- GTTAAGCAGGCTCCTCGGAA-3ʹ; and reverse, 5ʹ-TCCGCATACACCACAGACTC-3ʹ. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control and the primers were as follows: sense, 5ʹ-GTCAACGGATTTGGTCTGTATT-3ʹ; and reverse, 5ʹ-AGTCTTCTGGGTGGCAGTGAT-3ʹ.
Construction of Plasmid Vector
SPRY4-IT1 was synthesized and subcloned into the pLenti-GIII-CMV-GFP-2A-Puro vector. Ectopic expression of SPRY4-IT1 was achieved through pCDNA-SPRY4-IT1 transfection, with an empty pCDNA vector as a control. The expression levels of SPRY4-IT1 were detected by RT-qPCR.
Cell Counting Kit-8 (CCK8) Assay and Colony Formation Assay
The half-maximal inhibitory concentration (IC50) was determined using a CCK8 assay. Transfected cells were seeded in 96-well plates and maintained in standard growth medium. Overnight, cells were exposed to different concentrations of cisplatin. Cell viability was assessed at 0, 24, 48, 72 and 96 h with and without cisplatin treatment. CCK8 of 10 μL/well was added and incubated for another 2 h. Absorbance at 450 nm was then detected using an automatic microplate reader. For the colony formation assay, transfected cells (0.5x103 cells/well) were seeded into 6-well plates. After 14 days, cells were fixed with methanol and stained with 0.1% crystal violet, and the number of colonies was counted.
Flow Cytometric Analysis
A549/DDP control cells and treated cells were seeded in 6-well plates and then treated for 24 h in medium with or without DDP. The cells were then washed twice in cold PBS and fixed with 70% ethanol. After centrifugation at 2000 x g for 5 min, the cells were double stained with Annexin V-FITC and propidium iodide and analyzed using a flow cytometer equipped with CellQuest software.
Western Blot Assay
Cells were lyzed in RIPA buffer with a protease inhibitor cocktail and phenyl-methylsulfonyl fluoride. Protein lysates were separated via SDS-PAGE (10% gel) and transferred onto polyvinylidene fluoride membranes (EMD Millipore). Blocking was achieved with 5% skimmed milk for 1 h in Tris-buffered saline Tween 20, and the membrane was incubated with primary antibodies against E-cadherin (Proteintech, USA), Vimentin (Proteintech), MPZL-1 (Proteintech), and GAPDH (Proteintech), overnight at 4°C. The membrane was then incubated with secondary antibody for 1 h after washing with TBST three times. An anti-GAPDH antibody was used as a control. Signals were visualized using the ECL chromogenic substrate and quantified by densitometry using Quantity One software.
Tumor Formation Assay in a Nude Mouse Model
Ten female athymic BALB/c nude mice (4 weeks old) were maintained under pathogen-free conditions and randomly divided into two groups (5 for every group). A549/DDP cells were transiently transfected with pCDNA-SPRY4-IT1 and empty vector, and harvested from the 6-well cell culture plates, washed with phosphate-buffered saline, and resuspended at a concentration of 1x108 cells/mL. A total of 100 mL of suspended cells was subcutaneously injected into a single side of the posterior flank of each mouse. The tumor growth was examined every 3 days, and tumor volumes were calculated using the equation V=0.5xDxd2 (V, volume; D, longitudinal diameter; and d, latitudinal diameter). When tumor volumes reached 50 mm3, every mouse received cisplatin 3 times per week by intraperitoneal injection at a dose of 3.0 mg/kg. At 5 weeks post-injection, the mice were euthanized, and the subcutaneous growth of each tumor was examined. The present study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Nanjing Medical University.
Immunohistochemical (IHC)
The mouse xenograft tumors were immunostained for the marker of Ki-67, E-cadherin and Vimentin.
Statistical Analysis
All the experiments were replicated 3 times. All data were shown as means ± SD. Statistical differences were tested using Student’s t-test (two-tailed) between two groups and one-way analysis of variance (ANOVA) among multiple groups by SPSS 16.0 software. P≤0.05 was considered to be statistically significant.
Results
SPRY4-IT1 Is Downregulated in DDP-Resistant NSCLC Cells (A549/DDP)
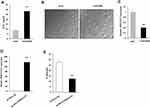
To evaluate the differences of A549/DDP and A549 cells on DDP treatment response, CCK8 assay was performed to detect the cell viability in the presence of DDP. It showed that A549/DPP cells were more resistant to DDP compared to A549 cells as determined by their IC50 (half-maximal inhibitory concentration) respectively (28.36± 2.3 µg/mL vs 8.75±1.0µg/mL; Figure 1A; P < 0.01). A549/DDP cell appears large swellings and spindle-shaped form different from A549 cell under microscopy (Figure 1B). Considering the role of SPRY4-IT1 in NSCLC development,1 we detected the expression of SPRY4-IT1 in these two cell lines. It was revealed that SPRY4-IT1 expression in A549/DDP cells was significantly decreased by approximately 2.5 folds (Figure 1C; P<0.01) in contrast to A549 cells. To further investigate the effects of SPRY4-IT1 on the development of DDP resistance in NSCLC, pCDNA-SPRY4-IT1 vector was transfected into A549/DDP cells to up-regulate SPRY4-IT1 expression (Figure 1D). CCK8 assay demonstrated that IC50 of SPRY4-IT1 over-expressed cells was largely decreased compared with controls (12.36±1.2µg/mL vs 27.44±2.5µg/mL; Figure 1E; P < 0.01).
SPRY4-IT1 Overexpression Inhibits Proliferation and Promotes Apoptosis in A549/DDP Cells
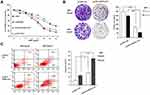
To investigate how SPRY4-IT1 influences the development of NSCLC cell resistance to DDP, the CCK8 assay was conducted. The results of CCK8 assay revealed that cell growth was impaired in A549/DDP cells transfected with pcDNA-SPRY4-IT1 combined with DDP treatment than that in controls (Figure 2A). To further verify the results, colony formation assay was performed and the results showed that clonogenic survival was inhibited following overexpression of SPRY4-IT1 under DDP treatment (DDP dose of 0.0, 8.0 μg/mL; Figure 2B). Furthermore, flow cytometry analysis showed that upregulation of SPRY4-IT1 expression significantly promoted DDP-induced apoptosis in A549/DDP cells (DDP dose of 0.0, 8.0 μg/mL; Figure 2C).
MPZL-1 Is Negatively Modulated by SPRY4-IT1 in A549/DDP Cells
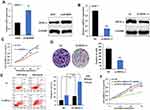
RNA transcriptome sequencing was applied to identify genes that exhibited altered expression between SPRY4-IT1 overexpressed cells and controls (Table 1). Ninety-two genes were differentially expressed in all the three replicates (expression change ≥1.5 fold; P<0.05), and 53 genes were upregulated and 39 genes were downregulated (Figure 3A). The Gene Ontology (Go) enrichment analysis demonstrated that those genes with significant differences (≥2.5 fold change) following SPRY4-IT1 overexpression were primarily involved in apoptosis, epithelial–mesenchymal transformation (EMT), and mesenchymal cell development pathways (Figure 3B). To validate the microarray findings, the top 10 up-regulated and down-regulated genes were subjected to verification by RT-qPCR (Figure 3C and Table 2). It was revealed that the trend in expression changes (upregulation or downregulation) of all 10 selected genes were consistent with the microarray analysis. However, among those differentiated-expression genes, MPZL-1 expression exhibited the most substantial change (down-regulated) and was enriched in the EMT-associated cell migration pathways. The results of the Western blot analysis further confirmed that upregulation of SPRY4-IT1 could lead to decreased MPZL-1 protein expression in A549/DDP cells (Figure 3D and E) while neither upregulation nor downregulation of MPZL-1 had no significant effect on the expression of SPRY4-IT1. These results indicated that the expression of MPZL-1 was negatively regulated by SPRY4-IT1.
 |
Table 1 Number of Regulated Genes in A549/DDP Cells After Overexpression of SPRY4-IT1 as Compared to Negative Controls |
 |
Table 2 The Dysregulated Gene Profiles After Overexpression of SPRY4-IT1 Using RNA Transcriptome Sequencing |
SPRY4-IT1 Reverses Cisplatin Resistance via Downregulating MPZL-1
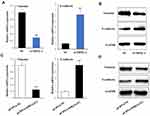
In order to investigate whether MPZL-1 participates in the development of chemoresistance in A549/DDP cells, we first detected the mRNA and protein expression levels of MPZL-1 in parental A549 and A549/DDP cells. It was confirmed that MPZL-1 expression was upregulated in A549/DDP cells compared with parental A549 cells significantly (Figure 4A; P<0.01). Next, it showed that decreased expression of MPZL-1 by siRNA-MPZL-1 transfection (Figure 4B) inhibited cell proliferation induced by cisplatin, as observed from the survival curve and clonogenic survival assay (Figure 4C and D). Furthermore, down-regulation of MPZL-1 promoted A549/DDP cell apoptosis (Figure 4E). To further verify whether MPZL-1 was involved in SPRY4-IT1-induced regulation of DDP sensitivity, we performed rescue experiments. A549/DDP cells were co-transfected with pCDNA-SPRY4-IT1 and pCDNA-MPZL-1, we found that over-expression of MPZL-1 partially compromised the inhibition effects of SPRY4-IT1 on growth of A549/DDP cells, thereby restoring DDP resistance (Figure 4F). Our results revealed that the effects of SPRY4-IT1 on NSCLC DDP resistance is at least in part through targeting MPZL-1.
SPRY4-IT1/MPZL-1 Affects Epithelial–Mesenchymal Transition in A549/DDP Cells
Our previous study has documented that SPRY4-IT1 could affect cell epithelial–mesenchymal transition (EMT) process in NSCLC cells, and EMT has been identified as an important effector in the development of chemoresistance. To verify whether SPRY4-IT1/MPZL-1 confer on NSCLC cells DDP resistance via affecting EMT, we detected E-cadherin and Vimentin mRNA and protein expression levels after up-regulation of SPRY4-IT1 or down-regulation of MPZL-1. The results of RT-qPCR (Figure 5A and C) and Western blot (Figure 5D and B) determined that over-expression of SPRY4-IT1 or down-regulation of MPZL-1 could increase E-cadherin expression while decrease Vimentin expression.
Overexpression of SPRY4-IT1 Inhibits Xenograft Tumor Growth Under DDP Treatment
In order to validate the effect of SPRY4-IT1 on the sensitivity of A549/DDP cells to DDP in vivo, we constructed nude mouse xenograft model. The results showed that the average tumor volume or weight of SPRT4-IT1 overexpression group was significantly smaller or lighter than that of controls under cisplatin treatment (Figure 6A–C). The expression of SPRY4-IT1 in tumor tissues from pCDNA-SPRY4-IT1 transfected A549/DDP cells was significantly increased than controls (Figure 6D). Moreover, immunohistochemical staining revealed decreased Ki-67 expression, reflecting proliferation inhibition in tumor tissues formed from A549/DDP cells with lncRNA SPRY4-IT1 overexpression (Figure 6E). In addition, relative expression of epithelial marker (E‑cadherin) was increased while mesenchymal marker (Vimentin) was decreased in pCDNA-SPRY4-IT1 group (Figure 6E).
Discussion
For NSCLC patients, chemotherapy remains to be the most basic and successful management; however, drug resistance, either primary or acquired, represents a severe barrier in the improvement of long-term survival and quality of life. With the development of molecular analytical and profiling techniques, the current understanding of resistance mechanisms and certain novel chemoresistance-associated lncRNAs have been identified.11 In the case of NSCLC, multiple lncRNAs, such as CCAT1, AK126698, HOTAIR, GAS5, UCA1 and MEG3 have been reported to influence chemoresistance.12 We previously reported that HOTAIR contributes to cisplatin resistance by downregulating P2113 and MEG3 has effect on mitochondrial apoptosis pathway to lead to cisplatin resistance.14 In this study, it was first revealed that lncRNA SPRY4-IT1 was downregulated in NSCLC cisplatin-resistant cells and up-regulation of SPRY4-IT1 partially reversed DDP resistance of NSCLC cells by promoting apoptosis and inhibiting proliferation.
Despite the detailed molecular mechanisms underlying chemoresistance not yet being fully understood, an accumulating researches have revealed the correlation of drug resistance and EMT.15,16 EMT is a physiopathological process in which epithelial cells lose adhesion proteins and acquire the characteristics of mesenchymal cells.17,18 According to certain clinical data regarding EGFR inhibitors (EGFR-TKIs), it has been suggested that EMT markers may be associated with limited response to EGFR inhibitors, whereas the retention of the epithelial phenotype is correlated with response, even in those without EGFR mutations. A study by Lim in breast cancer demonstrated that EMT confers chemoresistance by regulating the genes associated with cell death and stem cell maintenance. Another study by Pattabiraman revealed that abnormally activated EMT displayed elevated levels of ATP-binding cassette (ABC) that promoted drug efflux.21 In the present study, we observed that forced SPRY4-IT1 expression reversed the cisplatin-resistant phenotype of NSCLC, in large part, owing to its ability to inhibit EMT process.
Further RNA sequencing revealed that MPZL-1 may be a potential SPRY4-IT1 target in NSCLC DDP-resistant cells. In addition, MPZL-1 upregulation had been identified in NSCLC cisplatin-resistant cells (A549/DDP), and down-regulation of MPZL-1 reversed cisplatin resistance and suppressed EMT process. MPZL-1, also known as PZR, has been identified as Src homology phosphatase type-2 (SHP-2) binding partner in epithelial cells which is essential for haematopoietic skeletal and vascular development.19,20 Recent studies have shown that MPZL1 could promote the fibronectin-dependent migration of murine mesenchymal-derived MEF cells, and may be involved in adhesion-dependent signaling.21 Jia et al found that MPZL1 increased the in vitro migratory and in vivo metastatic potential of HCC cells by elevating phosphorylation of ERK-1/2 and AKT, which contribute to EMT programmes via receptor tyrosine kinase (RTK) pathway.22 So, we hypothesized that in NSCLC, MPZL-1 may act as an activator of intracellular signaling pathway regulating EMT, conferring a greater resistance to DDP of NSCLC cells. It has been established that lncRNAs can either act as positive or negative regulators of target gene expression, at various levels, including chromatin modification, transcription, and post-transcriptional, depending on their subcellular location.23,24 Various lncRNAs can recruit special chromatin remodeling complexes to particular chromatin loci to induce epigenetic silence.25 There are also reports on some lncRNAs which regulate the transcription of target genes via a direct mechanism,26 such as promoter and enhancer associated transcripts (paRNAs and eRNAs, respectively), besides, by regulating transcription of enhancers and promoters, they can play negative regulation of gene transcription. Additionally, the involvement of lncRNAs in post-transcriptional modifications can reflect gene function and stability. In summary, our findings first reveal that overexpression SPRY4-IT1 can reverse resistance to cisplatin of A549/DDP cells in vitro and vivo in lung cancers by downregulating MPZL-1 via suppressing EMT. However, the possible mechanisms underlying SPRY4-IT1 negative regulation of MPZL-1 remain to be discovered and need our further work.
Conclusions
SPRY4-IT1/MPZL-1 is a promising target for improving cisplatin efficiency in the treatment of NSCLC.
Acknowledgment
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81672307, 81501980, and 81871871), the Medical Innovation Team Foundation of the Jiangsu Provincial Enhancement Health Project (No. CXTDA2017021), the “333 high class Talented Man Project” (No. 2016-II-426), Jiangsu Provincial Six Talent Peaks (No. WSW-264), Education and Health Medicine Innovation Team Project (No. CXTDA2017042) and the Nanjing Science and Technology Development Project (No. 2017sc512046).
Disclosure
The authors have no conflicts of interest to declare in this work.
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi:10.3322/caac.v63.1
2. Salim H, Zong D, Hååg P, et al. DKK1 is a potential novel mediator of cisplatin-refractoriness in non-small cell lung cancer cell lines. BMC Cancer. 2015;15:628–643. doi:10.1186/s12885-015-1635-9
3. Gao Y, Liu Z, Zhang X, et al. Inhibition of cytoplasmic GSK-3β increases cisplatin resistance through activation of Wnt/β-catenin signaling in A549/DDP cells. Cancer Lett. 2013;336(1):231–239. doi:10.1016/j.canlet.2013.05.005
4. Tang L, Wang Y, Wang H, et al. Long noncoding-RNA component of mitochondrial RNA processing endoribonuclease is involved in the progression of cholangiocarcinoma by regulating microRNA-217. Cancer Sci. 2019;110(7):2166–2179. doi:10.1111/cas.2019.110.issue-7
5. Liu D, Li Y, Luo G, et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2017;388:281–291. doi:10.1016/j.canlet.2016.12.005
6. Zhang W, Lei P, Dong X, et al. The new concepts on overcoming drug resistance in lung cancer. Drug Des Devel Ther. 2014;8:735–744. doi:10.2147/DDDT.S60672
7. Khaitan D, Dinger ME, Mazar J, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71(11):3852–3862. doi:10.1158/0008-5472.CAN-10-4460
8. Shi Y, Li J, Liu Y, et al. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer. 2015;14:51. doi:10.1186/s12943-015-0318-0
9. Xie HW, Wu QQ, Zhu B, et al. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumor Biol. 2014;35:7743–7754. doi:10.1007/s13277-014-2013-y
10. Sun M, Liu X-H, Lu K-H, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial–mesenchymal transition. Cell Death Dis. 2014;5:e1298. doi:10.1038/cddis.2014.256
11. Xiong G, Feng M, Yang G, et al. The underlying mechanisms of non-coding RNAs in the chemoresistance of pancreatic cancer. Cancer Lett. 2017;397:94–102. doi:10.1016/j.canlet.2017.02.020
12. Wei L, Wang X, Lv L, et al. The emerging role of noncoding RNAs in colorectal cancer chemoresistance. [published online ahead of print July 29, 2019]. Cell Oncol (Dordr). 2019;42(6):757–768. doi:10.1007/s13402-019-00466-8
13. Liu Z, Sun M, Lu K, et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregulation of p21(WAF1/CIP1) expression. PLoS One. 2013;8(10):e77293. doi:10.1371/journal.pone.0077293
14. Liu J, Wan L, Lu K, et al. The noncoding RNA MEG3 contributes to cisplatin resistance of human lung adenocarcinoma. PLoS One. 2010;10(5):e0114586. doi:10.1371/journal.pone.0114586
15. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38–55. doi:10.1186/1476-4598-10-38
16. Zhang Q, Huang F, Yao Y, et al. Interaction of transforming growth factor-β-Smads/microRNA-362-3p/CD82 mediated by M2 macrophages promotes the process of epithelial-mesenchymal transition in hepatocellular carcinoma cells. Cancer Sci. 2019;110(8):2507–2519. doi:10.1111/cas.14101
17. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69–84. doi:10.1038/s41580-018-0080-4
18. Jiang JG, Ning YG, Chen C, et al. Cytochrome p450 epoxygenase promotes human cancer metastasis. Cancer Res. 2007;67(14):6665–6674. doi:10.1158/0008-5472.CAN-06-3643
19. Shi ZQ, Lu W, Feng GS. The Shp-2 tyrosine phosphatase has opposite effects in mediating the activation of extracellular signal-regulated and c-Jun-terminal mitogen-activated protein kinases. J Biol Chem. 1998;273(9):4904–4908. doi:10.1074/jbc.273.9.4904
20. Zannettino AC, Roubelakis M, Welldon KJ, et al. Novel mesenchymal and haematopoietic cell isoforms of the SHP-2 docking receptor, PZR: identification, molecular cloning and effects on cell migration. Biochem J. 2003;370(Pt2):537–549. doi:10.1042/bj20020935
21. Eminaga S, Bennett AM. Noonan syndrome-associated SHP-2/Ptpn11 mutants enhance SIRPalpha and PZR tyrosyl phosphorylation and promote adhesion-mediated ERK activation. J Biol Chem. 2008;283(22):15328–15338. doi:10.1074/jbc.M801382200
22. Jia D, Jing Y, Zhang Z, et al. Amplification of MPZL1/PZR promotes tumor cell migration through Src-mediated phosphorylation of cortactin in hepatocellular carcinoma. Cell Res. 2014;24(2):204–217. doi:10.1038/cr.2013.158
23. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi:10.1016/j.cell.2009.02.006
24. Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi:10.1126/science.1192002
25. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi:10.1038/nsmb.2480
26. Natoli G, Andrau J-C. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet. 2012;46:1–19. doi:10.1146/annurev-genet-110711-155459
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.