Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA SNHG14 Contributes to the Development of Hepatocellular Carcinoma via Sponging miR-217
Authors Xu X, Song F , Jiang X, Hong H, Fei Q, Jin Z, Zhu X, Dai B , Yang J, Sui C, Xu M
Received 2 January 2020
Accepted for publication 20 March 2020
Published 29 May 2020 Volume 2020:13 Pages 4865—4876
DOI https://doi.org/10.2147/OTT.S244530
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Xiaoyong Xu,1,* Feihong Song,2,* Xinwei Jiang,1,* Han Hong,1 Qiang Fei,1 Zhengkang Jin,1 Xiang Zhu,1 Binghua Dai,2 Jiamei Yang,2 Chengjun Sui,2 Minhui Xu1
1Department of Hepato-Pancreato-Biliary Surgery, The Affiliated Suzhou Hospital of Nanjing Medical University, Suzhou 215001, People’s Republic of China; 2Department of Special Treatment and Liver Transplantation, Shanghai Eastern Hepatobiliary Surgery Hospital, Shanghai 200438, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Minhui Xu
Department of Hepato-Pancreato-Biliary Surgery, The Affiliated Suzhou Hospital of Nanjing Medical University, Suzhou 215001, People’s Republic of China
Email [email protected]
Chengjun Sui
Department of Special Treatment and Liver Transplantation, Shanghai Eastern Hepatobiliary Surgery Hospital, Shanghai 200438, People’s Republic of China
Email [email protected]
Background: Thousands of long non-coding RNAs (lncRNAs) have been functionally verified as crucial regulators of physiological processes and disease progressions, yet their roles in hepatocellular carcinoma (HCC) have not been clearly illuminated.
Methods: We analyzed the expression of lncRNA-SNHG14 in TCGA data via bioinformatic analysis and detected its expression in HCC specimens by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Loss-of-function experiments were used to study the biological function of SNHG14 in HCC cells. RT-qPCR, Western blotting and dual-luciferase reporter assay were carried out to investigate the molecular mechanism of SNHG14 in HCC.
Results: The upregulation of lncRNA-SNHG14 was observed in HCC tissues compared with normal tissues via RT-qPCR and bioinformatic analysis of TCGA data. Silencing of SNHG14 inhibited cell proliferation and induced cell apoptosis in HCC cells. microRNA-217 (miR-217), the tumor-suppressive miRNA in HCC, was predicted and confirmed as a miRNA sponged by SNHG14 in HCC cells. Via downregulation of miR-217, SNHG14 increased the expression of several miR-217-related oncogenes and subsequently activated oncogene-related signaling pathways in HCC cells. In addition, inhibition of miR-217 reversed SNHG14 silencing induced decrease of cell proliferation and increase of cell apoptosis. Their association was verified in the published microarray dataset and the collected HCC samples.
Conclusion: In summary, SNHG14 is involved in the development of HCC via sponging miR-217 and it may be a biomarker for patients with HCC.
Keywords: SNHG14, hepatocellular carcinoma, miR-217
Introduction
Hepatocellular carcinoma (HCC) is the most prevalent cancer type of all liver cancer types.1 Due to the aggressive nature of HCC, the prognosis of patients with HCC is dismal, with the average overall survival of only approximately 10 months.2 It is pivotal to reveal the signaling network and provide novel targets for the diagnosis and treatment of patients with HCC.
Long non-coding RNAs (lncRNAs) are endogenous single-stranded RNA molecules with more than 200 nucleotides in length.3 LncRNAs can interact with RNA, protein and DNA in cells and participate in various biological processes.4 According to competing for endogenous RNA (ceRNA) hypothesis, lncRNAs may sponge microRNAs (miRNAs) to prevent miRNAs from binding to their target genes, thereby upregulating gene expression.5 Recent studies have suggested that several lncRNAs control the expression of key genes and the activity of key signaling pathways in HCC via the ceRNA mechanism.6–8 For example, LncRNA-LINC00346 promotes HCC development via sponging miR-542-3p, consequently activating the Wnt signaling in HCC cells.8 Mesenchymal stem cells facilitate epithelial–mesenchymal transition of HCC cells via a lncRNA-MUF/miR-34a/Snail1 axis.9 LncRNA-SNHG14 exhibits distinct functions in different cancer types.10,11 Most recently, Pu et al have reported the upregulation of SNHG14 in HCC, which promotes HCC progression via regulating miR-4673/SOCS1.12
The dysregulation of miRNAs is implicated in the initiation and progression of HCC.13 By targeting 3ʹUTR of target mRNAs, altered expression of miRNAs leads to the upregulation of oncogenes and downregulation of tumor suppressors in HCC cells.14,15 In recent years, numerous differentially expressed miRNAs have been identified in HCC tissues compared with normal tissues.16 Several miRNAs are confirmed as tumor-associated miRNAs via experimental validation.17,18 For instance, miR-217 is a well-characterized tumor-suppressive miRNA in HCC. Su et al firstly demonstrate that miR-217 is downregulated in metastatic HCC tissues and cells, in addition, they discover a miR-217/E2F3 axis in HCC cells.19 miR-217 expression is negatively correlated with metadherin (MTDH) and represses MTDH expression via direct binding to MTDH mRNA in HCC cells, thereby reducing cell proliferation, metastasis and inducing cell apoptosis.20 MiR-217 also suppresses cell proliferation and metastasis via targeting MAPK1.21 Analysis of miR-217 expression in HCC specimens suggests that decreased expression of miR-217 is associated with vascular invasion, advanced TNM stage and shorter overall survival.22 The downregulation of miR-217 in HCC is reported as a result of increased CRNDE and HOTAIR expression in HCC cells.21,23
In the present study, we aimed to further investigate the molecular mechanisms and biological functions of SNHG14 in HCC. We analyzed the expression of SNHG14 in HCC tissues and cells. Loss of function assays were performed to study the role of SNHG14 during HCC proliferation and survival. Bioinformatic analysis, dual-luciferase reporter assay, Western blotting and RT-qPCR were carried out to investigate the SNHG14/miR-217 interaction in HCC cells.
Materials and Methods
Collection of Tissue Samples
From June 2012 to September 2017, 55 pairs of HCC tissues and matched normal tissues were collected from patients with HCC in The Affiliated Suzhou Hospital of Nanjing Medical University. The study was performed in accordance with the Declaration of Helsinki. The protocol of experiments was approved by the Ethical Committee of The Affiliated Suzhou Hospital of Nanjing Medical University. Written informed consents were collected from all participants of the current study. All patients received no chemotherapy or radiotherapy before surgery. TNM staging was performed in accordance with the 2009 seventh edition of the UICC (Union for International Cancer Control). The samples were snap-freeze in liquid nitrogen then stored in −80°C refrigerator.
Cell Culture and Transfection
Transformed Human Liver Epithelial cell line (THLE-2) and the human hepatocellular carcinoma cell line (Huh-7, Hep3B) were bought from ATCC (Rockville, MD). Cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen; Thermo Fisher Scientific) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific). The cells were cultured at 37°C in a moist atmosphere with 5% CO2.
si-NC (Negative Control), si-SNHG14-1, si-SNHG14-2, miR-217-5p mimic, miR-NC mimic, miR-217-5p inhibitor and miR-NC inhibitor were synthesized by GenePharma (Suzhou, China). For silencing of SNHG14, si-NC or si-SNHG14-1 or si-SNHG14-2 were transfected into indicated cells with Lipofectamine RNAiMax (Invitrogen; Thermo Fisher Scientific) following manufacturer’s protocol. MiR-217 mimic, miR-NC mimic, miR-217 inhibitor or miR-NC inhibitor were transfected into cells with Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific). At 48 hours after transfection, cells were subjected to RNA extraction and RT-qPCR to validate transfection efficiency. The sequence: si-NC:5ʹ-CCAUGAGGUCAUGGUCUG-3ʹ; si-SNHG14-1:5ʹ-GAGAUGGAUCAACAGUAU-3ʹ; si-SNHG14-2:5ʹ-GCUACAAUCACUAUGAAUC-3ʹ; miR-217 mimic:5ʹ-UACUGCAUCAGGAACUGAUUGGA-3ʹ; miR-NC-mimic:5ʹ-UUGUCCGAACGUGUCACGU-3ʹ; miR-217 inhibitor:5ʹ-UCCAAUCAGUUCCUGAUGCAGUA-3ʹ; miR-NC inhibitor:5ʹ- CAGCUGGUUGAAGGGGACCAAA-3ʹ.
Reverse Transcription‐Quantitative Polymerase Chain Reaction (RT-qPCR)
RNA was extracted from cells and tissues by TRIzol reagent (Invitrogen; Thermo Fisher Scientific). RNA was reverse transcribed into cDNA with the RevertAid Reverse Transcriptase kit (Thermo Fisher Scientific). RT-qPCR was carried out with a PrimerScript one-step RT-PCR kit (TaKaRa, Tokyo, Japan). The relative gene expressions were calculated by the 2−ΔΔCt method24 with GAPDH and U6 snRNA serving as internal controls for mRNA and miRNA, respectively. Primer sequences: SNHG14-Forward: 5ʹ‐CAGGCTGAACTGAGGCAGGCAT‐3ʹ; SNHG14-Reverse: 5ʹ-ACATCTCATTCTATAGTCAATGT‐3ʹ; MTDH-Forward: 5ʹ‐AAATGGGCGGACTGTTGAAGT‐3ʹ; MTDH-Reverse: 5ʹ‐CTGTTTTGCACTGCTTTAGCAT‐3ʹ; KLF5-Forward: 5ʹ‐ CCTGGTCCAGACAAGATGTGA‐3ʹ; KLF5-Reverse: 5ʹ‐ GAACTGGTCTACGACTGAGGC‐3ʹ; E2F3-Forward: 5ʹ‐AGAAAGCGGTCATCAGTACCT‐3ʹ; E2F3-Reverse: 5ʹ‐TGGACTTCGTAGTGCAGCTCT‐3ʹ; IGF1R-Forward: 5ʹ‐ TCGACATCCGCAACGACTATC‐3ʹ; IGF1R-Reverse: 5ʹ‐ CCAGGGCGTAGTTGTAGAAGAG‐3ʹ; GAPDH-Forward: 5ʹ‐GAAGGTGAAGGTCGGAGTC‐3ʹ; GAPDH-Reverse: 5ʹ‐GAAGATGGTGATGGGATTTC‐3ʹ; Stem-loop primer:5ʹ-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCCAAT-3ʹ; miR-217-Forward:5ʹ-TCGGCAGGTACTGCATCAGGAA-3ʹ; miR-217-Reverse:5ʹ-CTCAACTGGTGTCGTGGA-3ʹ.
Western Blotting
ERK1/2 (4695, 1:2000), p-ERK1/2 (4370, 1:2000) and MTDH (14065, 1:2000) antibodies were purchased from Cell Signaling Technology (Carlsbad, CA). E2F3 (ab50917, 1:2000), KLF5 (ab137676, 1:2000) and GAPDH (ab8245, 1:10,000) antibodies were bought from Abcam (Cambridge, UK). HRP-conjugated secondary antibodies against rabbit (ab6721, 1:10,000) and mouse (ab205719, 1:10,000) were obtained from Abcam. Proteins were extracted from cells by RIPA lysis buffer (Thermo Fisher Scientific). Protein concentration was determined by a BCA Protein Assay kit (Thermo Fisher Scientific). A 25 μg proteins were loaded in 8% SDS-PAGE gels, transferred to PVDF membranes and blocked in 5% non-fat milk at room temperature for 1 hour. The membrane was incubated by primary antibodies and secondary antibodies at room temperature for 2 hours sequentially. The blots were developed with an ECL Western Blotting Substrate Kit (Pierce; Thermo Fisher Scientific). The intensity of bands was measured with Image J software 1.6.0 (National Institute of Science).
Dual-Luciferase Reporter Assay
Sequence of SNHG14 containing putative binding site (from 4000 bp to 5000 bp) for miR-217 was synthesized by GenePharma and ligated into pmirGLO plasmid (Promega, Hercules, CA). Two-point mutations were introduced to pmirGLO-SNHG14-WT to construct pmirGLO-SNHG14-Mut. Cells were transfected with SNHG14-WT or SNHG14-Mut in combination with miR-NC mimic or miR-217 mimic. At 48 hours after transfection, cells were harvested and the firefly luciferase and Renilla luciferase activities were determined with a Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase was normalized to Renilla luciferase in each group.
Bioinformatic Analysis
The interaction between miRNAs and lncRNA-SNHG14 was predicted by the miRDB database (http://mirdb.org/). The association between SNHG14 levels and IGF1R, E2F3, KLF5, and MTDH expression was analyzed by transcriptome data of The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) on the GEPIA (http://gepia.cancer-pku.cn/index.html). GEPIA was also used to explore the expression pattern of SNHG14 in hepatocellular carcinoma. Briefly, expression profile was constructed by selecting comparison of TCGA-LIHC (The Cancer Genome Atlas-Liver hepatocellular carcinoma) data (n=369) and TCGA normal data (n=50).
Flow Cytometry Analysis of Apoptosis
The percentage of apoptotic cells was detected with an Annexin V-FITC/PI Apoptosis Detection Kit (Invitrogen). In a brief, cells were harvested, suspended in Annexin binding buffer (from kit) and incubated with PI and Annexin V for 30 mins at room temperature. The cells were subjected to flow cytometry analysis. PI+/Annexin V+ cells were considered as later apoptotic cells, PI-/Annexin V+ cells were early apoptotic cells and PI-/Annexin V cells were viable cells.
Determination of Cell Proliferative Ability
The transfected cells were plated in each well of 96-well plates and incubated for 0, 24, 48, and 72 h, respectively. In each time point, 10 μL CCK-8 solution was added into indicated well and cultured for 2 hours at 37°C. The medium was transferred to wells in a new 96-well plate. The absorbance at 450 nM was detected to manifest cell proliferative ability.
Statistical Analysis
All data were analyzed by Graphpad Prism 5.0 and presented as mean±SD. Groups were compared with Student’s t test or one-way ANOVA followed by Tukey’s test. The association between clinicopathological parameters and SNHG14 expression was analyzed by Chi-square test. All experiments were repeated three times. P value of less than 0.05 was considered as statistically significant.
Results
SNHG14 Expression Was Increased in HCC Tissues
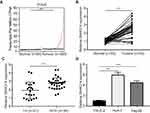
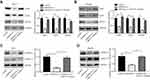
To study the potential role of lncRNA-SNHG14 in HCC, we analyzed SNHG14 expression in 369 HCC tissues and 50 normal liver tissues via bioinformatic analysis of TCGA-LIHC and TCGA normal liver tissues data using GEPIA software. The level of SNHG14 was higher in HCC tissues compared with normal liver tissues (Figure 1A). For validation, we collected 55 pairs of HCC tissues and matched normal tissues from patients and detected SNHG14 expression by RT-qPCR. Consistently, there was a significant elevation of SNHG14 expression in HCC tissues than normal tissues (Figure 1B). Furthermore, higher expression of SNHG14 was associated with later stage HCC (Stage III–IV) (Figure 1C). The expression of SNHG14 was not associated with tumor size, gender, age, AFP concentration, HBsAg status of HCC patients (Table 1). Furthermore, we detected SNHG14 expression in a panel of cell lines including HCC cell lines (Huh-7, Hep3B) and normal liver epithelial cell line THLE-2. It was observed that SNHG14 was significantly upregulated in Huh-7 and Hep3B in comparison with THLE-2 (Figure 1D).
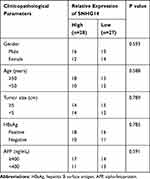 |
Table 1 The Association Between SNHG14 Expression and Clinicopathological Parameters in 55 Patients with Hepatocellular Carcinoma |
Knockdown of SNHG14 Suppressed HCC Cell Proliferation and Induced Cell Apoptosis
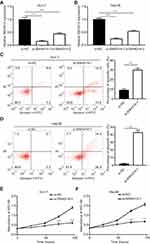
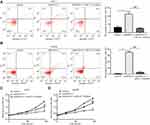
To determine the biological role of SNHG14 in HCC, siRNAs targeting SNHG14 was transfected into two HCC cell lines, Huh-7 and Hep3B. Transfection of two independent siRNAs of SNHG14 decreased SNHG14 expression in these two cell lines with the knockdown efficiency of around 85% and 50%, respectively (Figure 2A and B). Due to the relatively higher efficiency of si-SNHG14-1 than si-SNHG14-2, we chose it for further study. Knockdown of SNHG14 induced a significant elevation of apoptotic cells in Huh-7 (10% vs 30%) and Hep3B (0.5% vs 40%) cells (Figure 2C and D). Additionally, knockdown of SNHG14 caused a significant decrease in cell proliferation in Huh-7 and Hep3B cells, as measured by CCK-8 assay (Figure 2E and F). These data demonstrated that SNHG14 was pivotal for cell proliferation and resistance to apoptosis in HCC cells.
SNHG14 Repressed miR-217-5p Expression via Directly Sponging miR-217-5p in HCC Cells
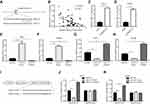
For further investigations on molecular mechanisms, miRDB software was used to predict the potential targets of SNHG14. Through bioinformatic analysis and literature review, we noticed that there was a putative binding site for miR-217, a known tumor suppressor, on the sequence of SNHG14 (Figure 3A). Thereafter, we detected miR-217 expression in 55 pairs of HCC tissues and matched normal tissues. Pearson correlation analysis suggested that there was a negative correlation between miR-217 and SNHG14 levels in 55 HCC tissues (r=−0.4369, p<0.001) (Figure 3B). Knockdown of SNHG14 increased miR-217 level in Huh-7 and Hep3B cells (Figure 3C and D). To examine the effect of miR-217 on SNHG14 expression, miR-217 mimic or miR-217 inhibitor was transfected into HCC cells to manipulate miR-217 expression. MiR-217 mimic increased miR-217 expression while miR-217 inhibitor decreased miR-217 expression in Huh-7 and Hep3B cells (Figure 3E and F). Overexpression of miR-217 decreased SNHG14 expression while miR-217 inhibition increased SNHG14 expression in Huh-7 and Hep3B cells (Figure 3G and H).
We next constructed SNHG14 luciferase plasmid with SNHG14-WT or SNHG14-Mut to further verify the relationship between SNHG14 and miR-217 in HCC (Figure 3I). Dual-luciferase reporter assay data showed that miR-217 overexpression decreased luciferase activity of SNHG14-WT, and miR-217 inhibition increased luciferase activity of SNHG14-WT, whereas the luciferase activity of SNHG14-Mut was not affected by miR-217 mimic or miR-217 inhibitor in Huh-7 cells (Figure 3J). Similar results were observed in Hep3B cells (Figure 3K). The data indicated that SNHG14 sponged miR-217 in HCC cells.
SNHG14 Upregulated Multiple Oncogenes in HCC Cells via Sponging miR-217
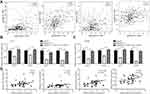
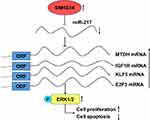
As a tumor-suppressive miRNA, miR-217 targeted several oncogenes in HCC cells to repress cell proliferation and induce cell apoptosis.19,20,25,26 Via bioinformatic analysis of transcriptome data of TCGA-LIHC, it was found that SNHG14 expression was positively correlated with the reported miR-217 target genes in HCC, including IGF1R, E2F3, KLF5 and MTDH in 369 HCC tissues (Figure 4A). We performed RT-qPCR to detect the expression of miR-217 target genes following SNHG14 knockdown with or without miR-217 inhibition in HCC cells. SNHG14 knockdown reduced IGF1R, E2F3, KLF5 and MTDH mRNA expression in Huh-7 cells which was reversed upon miR-217 inhibition (Figure 4B). Similarly, mRNA levels of miR-217 target genes were downregulated after SNHG14 silencing which was reversed upon miR-217 inhibition in Hep3B cells (Figure 4C). More importantly, in our collected tumor samples, it was observed that SNHG14 expression was positively correlated with mRNA levels of IGF1R, E2F3, KLF5 and MTDH (Figure 4D), indicating their association was clinically relevant.
We also detected protein levels of these genes in HCC cells. In consistent with RT-qPCR data, SNHG14 knockdown decreased KLF5, E2F3 and MTDH protein expression in Huh7 cells and miR-217 inhibitor abrogated the downregulation of CHEK1, E2F3 and MTDH protein expression led by SNHG14 silencing (Figure 5A). Similar results were observed in Hep3B cells (Figure 5B). As acknowledged, MTDH facilitated HCC progression via activating MAPK/ERK signaling.27 Western blotting showed that SNHG14 knockdown reduced the ratio of phosphorylated ERK1/2 (p-ERK1/2) to total ERK1/2 (t-ERK1/2) in Huh-7 cells (Figure 5C), indicating SNHG14 positively regulated MAPK/ERK signaling. The results were also observed in Hep3B cells (Figure 5D). Collectively, the data suggested that SNHG14 promoted the expression of several key oncogenes via sponging miR-217 in HCC cells.
The Effect of SNHG14 Knockdown on HCC Cells Was Attenuated by miR-217 Inhibition
We next studied whether SNHG14 regulated HCC cell proliferation and apoptosis via sponging miR-217. In Huh-7 cells, inhibition of miR-217 attenuated an increased percentage of apoptotic cells induced by SNHG14 silencing (Figure 6A). Similar results were observed in Hep3B cells (Figure 6B). In the CCK-8 assay, it was found that miR-217 inhibitor could reverse the inhibition of cell proliferation induced by SNHG14 silencing in both Huh-7 and Hep3B cells (Figure 6C and D). The results suggested that SNHG14 controlled HCC progression via sponging miR-217.
Discussion
SNHG14 is an oncogenic lncRNA in several cancer types via sponging tumor-suppressive miRNAs.28–30 For example, SNHG14 promoted cell migration, invasion, proliferation and resistance to cell apoptosis in gastric cancer via sponging miR-145 and upregulation of SOX9.29 Most recently, Pu et al reported the aberrant expression of SNHG14 in HCC through bioinformatics analysis and found that SNHG14 promoted cell proliferation and inhibited cell apoptosis in Huh-7 cells partially via sponging miR-4673 and upregulation of SOCS1.12 In the present study, we confirmed the upregulation of SNHG14 in collected HCC specimens. Our data additionally suggested that SNHG14 expression was associated with HCC stage. Similar to the pro-proliferation and anti-apoptosis function of SNHG14 in gastric cancer and non-small cell lung cancer,10,29 we found that SNHG14 also promoted cell proliferation and resistance to apoptosis in HCC. The data demonstrated an oncogenic potential of SNHG14 in HCC.
Low expression of miR-217 was associated with vascular invasion, advanced TNM stage and poor prognosis in patients with HCC.22 miR-217 was then discovered as a decreased miRNA in HCC cells with strong invasive potential.19 The study found that miR-217 inhibited cell migration and invasion in HCC cells via targeting transcription factor E2F3.19 Despite its involvement in metastasis, miR-217 also suppressed cell proliferation and promoted cell apoptosis via targeting MTDH in HCC cells.20 miR-217 was also a downregulated miRNA in ovarian cancer and triple-negative breast cancer, and miR-217 directly targeted IGF1R and KLF5 to inhibit cancer cell proliferation.25,26 Previous studies suggested that miR-217 was sponged by several lncRNAs in cancer cells, including HOTAIR, MALAT1 and CRNDE.21,31,32 We found that miR-217 was directly sponged by SNHG14 in HCC cells and miR-217 expression was negatively correlated with SNHG14 in HCC tissues. Furthermore, SNHG14 expression was positively correlated with E2F3, IGF1R, MTDH and KLF5 levels in TCGA-LIHC data. RT-qPCR and Western blotting verified that SNHG14 regulated these oncogenes via sponging miR-217. IGF1R and MTDH were two well-known drivers of carcinogenesis in liver via activating MAPK/ERK signaling.27,33 We observed that the activity of MAPK/ERK signaling is controlled by SNHG14/miR-217 in HCC cells, indicating a pivotal role of SNHG14/miR-217 during HCC progression. Most importantly, inhibition of miR-217 could reverse the biological function of SNHG14 silencing in HCC cells. These data suggested a SNHG14/miR-217 axis in mediating HCC cell proliferation and resistance to cell apoptosis.
Conclusion
In conclusion, we demonstrated that SNHG14 was an upregulated lncRNA in HCC. SNHG14 facilitated cell proliferation and survival of HCC cells via sponging miR-217 and upregulation of several key oncogenes (IGF1R, KLF5, E2F3, MTDH) (Figure 7). The results manifested that SNHG14 was a potential biomarker for patients with HCC.
Acknowledgments
This study was supported by Project of Science and Technology Development Plan in Suzhou (SYSD2018138), Project of Clinical Expert Team Introduction in Suzhou (SZYJTD201807) and Natural Science Foundation of Shanghai (17ZR1438000).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Chan SC. Liver transplantation for hepatocellular carcinoma. Liver Cancer. 2013;2(3–4):338–344. doi:10.1159/000343849
3. Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108(10):1927–1933. doi:10.1111/cas.13342
4. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
5. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
6. Wang Y, He L, Du Y, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413–425. doi:10.1016/j.stem.2015.03.003
7. Wang X, Sun W, Shen W, et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J Hepatol. 2016;64(6):1283–1294. doi:10.1016/j.jhep.2016.01.019
8. Zhang N, Chen X. A positive feedback loop involving the LINC00346/beta-catenin/MYC axis promotes hepatocellular carcinoma development. Cell Oncol (Dordr). 2019.
9. Yan X, Zhang D, Wu W, et al. Mesenchymal stem cells promote hepatocarcinogenesis via lncRNA-MUF interaction with ANXA2 and miR-34a. Cancer Res. 2017;77(23):6704–6716. doi:10.1158/0008-5472.CAN-17-1915
10. Jiao P, Hou J, Yao M, et al. SNHG14 silencing suppresses the progression and promotes cisplatin sensitivity in non-small cell lung cancer. Biomed Pharmacother. 2019;117:109164. doi:10.1016/j.biopha.2019.109164
11. Zhang W, Duan W, Mo Z, et al. Upregulation of SNHG14 suppresses cell proliferation and metastasis of colorectal cancer by targeting miR-92b-3p. J Cell Biochem. 2019.
12. Pu J, Wei H, Tan C, et al. Long noncoding RNA SNHG14 facilitates hepatocellular carcinoma progression through regulating miR-4673/SOCS1. Am J Transl Res. 2019;11(9):5897–5904.
13. Klingenberg M, Matsuda A, Diederichs S, et al. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67(3):603–618. doi:10.1016/j.jhep.2017.04.009
14. Yang Y, Zhao Z, Hou N, et al. MicroRNA214 targets Wnt3a to suppress liver cancer cell proliferation. Mol Med Rep. 2017;16(5):6920–6927. doi:10.3892/mmr.2017.7483
15. Zhang Y, Huang F, Wang J, et al. MiR-15b mediates liver cancer cells proliferation through targeting BCL-2. Int J Clin Exp Pathol. 2015;8(12):15677–15683.
16. Niu JX, Meng XK, Ren JJ. Studied microRNA gene expression in human hepatocellular carcinoma by microRNA microarray techniques. World J Gastroenterol. 2015;21(44):12605–12611. doi:10.3748/wjg.v21.i44.12605
17. Feng Q, Zhang H, Nie X, et al. miRNA-149* suppresses liver cancer progression by down-regulating TRADD protein expression. Am J Pathol. 2019;190(2):469–483. doi:10.1016/j.ajpath.2019.10.010
18. Tao C, Sun H, Sang W, et al. miRNA-99a inhibits cell invasion and migration in liver cancer by directly targeting HOXA1. Oncol Lett. 2019;17(6):5108–5114. doi:10.3892/ol.2019.10199
19. Su J, Wang Q, Liu Y, et al. miR-217 inhibits invasion of hepatocellular carcinoma cells through direct suppression of E2F3. Mol Cell Biochem. 2014;392(1–2):289–296. doi:10.1007/s11010-014-2039-x
20. Zhang M, Li M, Li N, et al. miR-217 suppresses proliferation, migration, and invasion promoting apoptosis via targeting MTDH in hepatocellular carcinoma. Oncol Rep. 2017;37(3):1772–1778. doi:10.3892/or.2017.5401
21. Wang H, Ke J, Guo Q, et al. Long non-coding RNA CRNDE promotes the proliferation, migration and invasion of hepatocellular carcinoma cells through miR-217/MAPK1 axis. J Cell Mol Med. 2018;22(12):5862–5876. doi:10.1111/jcmm.13856
22. Tian YW, Shen Q, Jiang QF, et al. Decreased levels of miR-34a and miR-217 act as predictor biomarkers of aggressive progression and poor prognosis in hepatocellular carcinoma. Minerva Med. 2017;108(2):108–113. doi:10.23736/S0026-4806.16.04616-4
23. Wang LP, Wang JP, Wang XP. HOTAIR contributes to the growth of liver cancer via targeting miR-217. Oncol Lett. 2018;15(5):7963–7972. doi:10.3892/ol.2018.8341
24. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
25. Li J, Li D, Zhang W. Tumor suppressor role of miR-217 in human epithelial ovarian cancer by targeting IGF1R. Oncol Rep. 2016;35(3):1671–1679. doi:10.3892/or.2015.4498
26. Zhou W, Song F, Wu Q, et al. miR-217 inhibits triple-negative breast cancer cell growth, migration, and invasion through targeting KLF5. PLoS One. 2017;12(4):e0176395. doi:10.1371/journal.pone.0176395
27. Yoo BK, Emdad L, Su ZZ, et al. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest. 2009;119(3):465–477. doi:10.1172/JCI36460
28. Qi X, Shao M, Sun H, et al. Long non-coding RNA SNHG14 promotes microglia activation by regulating miR-145-5p/PLA2G4A in cerebral infarction. Neuroscience. 2017;348:98–106. doi:10.1016/j.neuroscience.2017.02.002
29. Liu Z, Yan Y, Cao S, et al. Long non-coding RNA SNHG14 contributes to gastric cancer development through targeting miR-145/SOX9 axis. J Cell Biochem. 2018;119(8):6905–6913. doi:10.1002/jcb.26889
30. Zhang YY, Li M, Xu YD, et al. LncRNA SNHG14 promotes the development of cervical cancer and predicts poor prognosis. Eur Rev Med Pharmacol Sci. 2019;23(9):3664–3671. doi:10.26355/eurrev_201905_17790
31. Hong Q, Li O, Zheng W, et al. LncRNA HOTAIR regulates HIF-1alpha/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis. 2017;8(5):e2772. doi:10.1038/cddis.2017.181
32. Liu P, Yang H, Zhang J, et al. The lncRNA MALAT1 acts as a competing endogenous RNA to regulate KRAS expression by sponging miR-217 in pancreatic ductal adenocarcinoma. Sci Rep. 2017;7(1):5186. doi:10.1038/s41598-017-05274-4
33. D’Alessandro R, Refolo MG, Lippolis C, et al. Strong enhancement by IGF1-R antagonists of hepatocellular carcinoma cell migration inhibition by Sorafenib and/or vitamin K1. Cell Oncol (Dordr). 2018;41(3):283–296. doi:10.1007/s13402-018-0370-z
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.