Back to Journals » Cancer Management and Research » Volume 11
Long Non-Coding RNA LINC00511 Mediates the Effects of ESR1 on Proliferation and Invasion of Ovarian Cancer Through miR-424-5p and miR-370-5p
Authors Wang K, Zhu G, Bao S, Chen S
Received 23 September 2019
Accepted for publication 23 November 2019
Published 27 December 2019 Volume 2019:11 Pages 10807—10819
DOI https://doi.org/10.2147/CMAR.S232140
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Kang Wang,1,2 Genhai Zhu,2 Shan Bao,2 Shiling Chen1
1Department of Gynecology and Obstetrics, Center for Reproductive Medicine, Nanfang Hospital, Southern Medical University, Guangzhou 510515, People’s Republic of China; 2Department of Gynecology, Hainan General Hospital, Haikou, Hainan 570000, People’s Republic of China
Correspondence: Shiling Chen
Department of Gynecology and Obstetrics, Center for Reproductive Medicine, Nanfang Hospital, Southern Medical University, Guangzhou 510515, People’s Republic of China
Email [email protected]
Introduction: Estrogen receptor 1 (ESR1) plays an important role in the pathological events of ovarian cancer (OV), but the underlying mechanism is not completely understood. Using bioinformatics analysis, we found that ESR1 is involved in the regulation of some lncRNAs that are highly expressed in ovarian cancer. The lncRNAs might mediate the roles of ESR1 in OV occurrence and progression.
Methods: This study measured the expression of the lncRNAs in OV cell lines using qRT-PCR. Some of the lncRNAs were silenced or overexpressed to determine their effects on the growth and invasion of CAOV3 cells with the stimulation of 17 beta-estradiol or not.
Results: ESR1-expressing OV cells (CAOV3 cells) shows higher LINC00511 and RP11-166P13.3 expression than the ESR1-losing OV cells (UWB1.289 cells). Depletion of the two lncRNAs enhanced cell viability and invasion and decreased apoptosis rate. In these respects, effects of LINC00511 were more remarkable than that those of RP11-166P13.3. Treatment with 17 beta-estradiol to stimulate ESR1 increased LINC00511 expression, while ESR1 inhibitor Fulvestrant decreased LINC00511 expression. FISH assay confirmed that LINC00511 is present in the cytoplasm and nucleus. Bioinformatics analysis revealed the interaction of LINC00511 with miR-424-5p and miR-370-5p, which was further identified by RNA-pull down assay. As indicated by RIP assay, silencing LINC00511 increased the interaction between Ago protein and these two miRNAs.
Discussion: Our study showed that ESR1-induced upregulation of LINC00511 promoted proliferation and invasion of CAOV3 cells probably through sponging miR-424-5p and miR-370-5p.
Keywords: ESR1, long non-coding RNA LINC00511, progression, ovarian cancer
Introduction
Ovarian cancer (OV) is one of the most common cancers and the fifth lethal gynecological cancer in women.1 It was estimated that there are 1,762,450 new cases and 606,880 deaths from cancers in 2019 in the United States.2 There will be 22,530 new cases and 13,980 deaths from ovarian cancer.2 Despite great progress in surgery and chemotherapy, the five-year survival rate for this patient population remains low, especially in patients with peritoneal metastases. Ovary belongs to gonadal tissue. OV is considered as a hormone-dependent cancer with the expression of estrogen receptors (ERs) and progesterone receptors (PRs) in 60–100% OV patients.3 Therefore, it is essential to clarify the pathological mechanisms and explored novel avenues to fight against this fatal cancer (including hormonal therapy).
Cellular signaling of estrogens is mediated through two ERs, ERα (a classical estrogen receptor subtype) and ERβ.4,5 Estrogen-bound ERα plays a pivotal role in follicle maturity, release and implantation. As ovarian cancer is a hormone-dependent cancer, imbalance of ER/PR is associated with a greater risk of ovarian carcinoma.6 ESR1 gene encodes ERα, a ligand-activated transcription factor, which is composed of several domains important for hormone binding, DNA binding, and activation of transcription ERα localizes to the nucleus where it may form a homodimer or a heterodimer with estrogen receptor 2. A large number of studies showed that Estrogen receptors are involved in various cancers such as OV, breast cancer, and endometrial cancer,6–8 but the underlying mechanism is not completely understood.
Long noncoding RNAs (lncRNAs) is a type of non-coding RNA with the length >200nt.9 They play an important role in human biological process and pathophysiology,10 and some of them have been found to participate in tumorigenesis.11 The molecular mechanisms of LncRNAs are varied, including acting as scaffolds, decoys, signals transducer and regulation in cis or trans.10–13 For example, PVT1 promotes the proliferation of OV by increasing SOX2.12 LINC00460 accelerates OV progression by the sponging effect on miRNA-338-3p.13
In this experiment, we found that ESR1 was involved in the transcriptional regulation of several lncRNAs. Among them, LINC00511 was closely linked with the proliferation and invasion of OV cells. Further study found that LINC00511 located in cytoplasm and nucleus and inhibited the expression of a series of miRNAs including miR-9-5p, miR-424-5p, and miR-183-5p. All these miRNAs have reported to suppress OV proliferation and invasion. Therefore, this study hence added further understanding of the cancer-promoting effect of ESR1 by up-regulating LINC00511.
Materials and Methods
Cell Lines Culture and Treatment
ESR1-expressing OV cell lines, including CAOV3, OVCAR3 and SKOV3, as well as ESR1-losing OV cell line (UWB1.289) were used in this study. These cell lines were obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China). Cells were cultured in DMEM (Gibco, CA, USA) containing 10% FBS (Gibco, CA, USA), 100 units/mL of penicillin–streptomycin (Invitrogen, CA, USA) in a humidified incubator at 37°C with 5% CO2. Estrogen receptor activator, receptor-17 β-estradiol (17β-E2, 10 nM estradiol, Sigma-Aldrich), and inhibitor, Fulvestrant (0.094 nM, Sigma-Aldrich) were added to CAOV-3 cells to stimulate and block ESR1 functions, respectively.
RNA Extraction and Quantitative Polymerase Chain Reaction (qPCR) Analysis
Total RNA was isolated from CAOV3, OVCAR3, SKOV3 and UWB1.289 cells, using the Trizol reagent (Takara, Dalian, China). The real-time PCR reactions were performed in triplicate and the relative expression levels of genes were analyzed by using the 2−ΔΔCt method. U6 and GAPDH were employed as miRNA and lncRNA internal control. The qRT-PCR reaction was performed in the ABI 7500 real-time PCR system (Applied Biosystems, CA, USA). The primer sequences are presented in Table 1.
 |
Table 1 The Sequence of Primers Used in PCR |
Western Blot Analysis
Cells were lysed in buffer containing 50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1% (w/v) CHAPS and protease inhibitor cocktail (Sigma), and the total cell lysates were resolved with SDS-PAGE gels. The following antibodies were used: ESR1 (Abcam, ab108398), GAPDH (Abcam, ab125247). HRP-linked secondary antibodies were used and blots visualized with the ECL kit (Millipore Corporation, Billerica, WBLUR0100).
Transfection
The small hairpin RNA (shRNAs) specifically targeting LINC00511 and RP11-166P13.3 were synthesized by GenePharma (Shanghai, China). The shRNAs were inserted into a lentiviral plasmid, pGLVU6/GFP (GenePharma). The shRNA-pGLVU6/GFP was transfected into CAOV3 cells using the Lipofectamine 2000 kit (Invitrogen) according to the manufacturer’s instructions. For ectopic expression of LINC00511 and RP11-166P13.3 in CAOV3 cells, transfection of pEGFP-C1-LINC00511 and pEGFP-C1-RP11-166P13.3 vectors (Genephama Biotech) was performed using the Lipofectamine 2000 (Invitrogen), according to the company’s instructions. The transfected cells were harvested at 12, 24, 48, or 72 hrs after transfection.
Cell Viability Assay
Cell viability was assessed by MTT [3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] cell viability. The cells (1×103) were seeded into 96-well plates. The absorbance was measured using an automatic microplate reader (Tecan, NANOQUANT, Switzerland). The experiment was repeated three times.
Cell Apoptosis Assay
CAOV-3 cells were harvested and washed with cold phosphate-buffered saline (PBS), then stained with 5 μL of AnnexinV-FITC (KeyGen Biotech, Shanghai, China) and 10 μL of PI (BD Pharmingen, San Diego, CA, USA) in dark. After that, the number of apoptotic cells were measured by flow cytometry (BD Biosciences, San Jose, CA, USA).
Trans-Well Assays
CAOV3 cells were cultured in medium without serum in the upper chamber of a trans-well (24-well insert, 8-mm pore size, Millipore). The upper chamber was coated with Matrigel (Sigma-Aldrich; Merck KGaA). Complete medium was subsequently added to the lower chamber. Following culture for 24 h, non-invading cells on the upper surface of the membrane were removed with a cotton-tipped swab, while the invading cells on the surface of the lower chamber were stained with 0.1% crystal violet at room temperature for 20 min. The invading cells were quantified by counting 10 random fields of view at x200 magnification using a light microscope (E200; Nikon Corporation, Tokyo, Japan).
Wound-Healing Assay
Wound healing assay was used to investigate cell migration. CAOV3 cells (5 × 105) were seeded in a six-well plate and then treated with siRNA-LINC00511, 17β-E2 or Fulvestrant. A 1-mL pipette tip was used to aggregate a “scratch-wound” on the cell monolayer. The culture medium was then replaced with FBS-free medium. Microscope images of the cells were captured immediately following scratching and following 24 h. The cell migration rate was calculated based on the movement of cells from initial placement to the final distance travelled following 24 h. Cell migration rate was calculated using the following equation: (initial distance – final distance)/initial distance x100.
Chromatin Immunoprecipitation (ChIP) Assay
CAOV3 cells were fixed using 1% formaldehyde and harvested on ice with ChIP lysis buffer (50 mM Tris-HCl ph 8.0, 5 mM EDTA, 0.1% deoxycholate, 1% Triton X-100, 150 mM NaCl and proteinase inhibitor). Subsequently, cells were sonicated and the supernatant was collected and incubated with dynabeads protein G and anti-ESR1 antibody. IgG was used as a negative control. After 2 h, the complex was washed three times and DNA was purified and condensed. Finally, the DNA fraction was analyzed by qRT-PCR.
Fluorescence in situ Hybridization (FISH)
FISH assay was employed to determine the localization of LINC00511 in CAOV3 cells. The LINC00511 probe was purchased from RiboBio, Co., Ltd. (Guangzhou China). CAOV3 cells were incubated with LINC00511 probe overnight at 37 °C. Then, the cells were washed and blocked by 3% BSA. After the cells were washed with Phosphate-Buffered Saline/Tween (PBST) three times, the nucleus was stained using 4′,6-Diamidino-2-Phenylindole (DAPI) diluted by PBST at the ratio of 1:800. The images of cells were captured under 5 different visual fields using the fluorescence microscope (Olympus Optical Co., Ltd., Tokyo, Japan).
RNA Pull-Down Assay
Biotinylated RNA probes including Bio-miR-NC, Bio-miR-424-5p, Bio-miR-424-5p Mut, Bio-miR-370-5p and Bio-miR-370-5p Mut were synthesized by KeyGen Biotech Company. These RNA probes were incubated with the lysates of CAOV3 cells and extracted using streptavidin-coupled magnetic beads according to the instructions of Pierce™ Magnetic RNA Pull-Down Kit (Rockford, IL, USA). RNA-RNA complexes were then eluted using the salt solution, and purified using TRIzol (Pierce). The enrichment of LINC00511 in the RNA-RNA complexes was quantified using qPCR, as described above.
RNA Immunoprecipitation (RIP)
RIP was performed on CAOV3 cells using EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore). Briefly, CAOV3 cells were lysed with RIPA-2 buffer and then incubated with Protein-A Dynabeads conjugated with IgG or Ago antibodies. The bead/antibody/lysate mixture was incubated at 4°C overnight rotating end-over-end. Beads were washed with cold NT2 buffer five times. Proteinase K treatment released RNAs from bound proteins and input and bound RNA was isolated with TRIzol (Invitrogen) and reverse transcribed as described above. Enrichment for miR-424-5p and miR-370-5p with Ago immunoprecipitation was calculated using the comparative Ct method, with samples normalized to input and compared to IgG control. Data are presented as fold enrichment relative to GAPDH enrichment for each sample.
Tumor Xenograft in Nude Mice
The animal experiment was approved by the Ethical Committee for Animal Research of Southern Medical University (protocol number: 2011-020) and conducted according to the National Institutes of Health guidelines for animal welfare. Nude mice (4–5 weeks old, male) were purchased from the Central Animal Facility of Southern Medical University. Two hundred milliliters of CAOV3 cells (1×106) were injected into the left side on the back of each mouse. The tumor volume was monitored on day 16. The tumor volume was calculated as v=a×b2/2.
Immunohistochemistry
Formalin-fixed and paraffin-embedded tissues of the xenografted CAOV3 tumor were deparaffinized and rehydrated. Tissues were treated for 20 min at 100°C in an autoclave for antigen retrieval and blocked with a blocking reagent (Protein Block Serum-Free, Dako Cytomation, Glostrup, Denmark) to avoid non-specific reactions. Then, tissues in sections were incubated with anti-Ki67 antibody (Dilution 1:400, ab16667, Abcam) overnight at 4°C, followed by horseradish peroxidase (HRP)-labeled anti-rabbit IgG (Histofine, Simple stain MAX-PO; Nichirei, Tokyo, Japan) for 30 min at room temperature. The sections were treated with 3, 3′-diaminobenzidine tetrahydrochloride solution.
Statistical Analysis
Statistical analysis was performed with the Student unpaired t-test. Statistical analysis of the data was performed using SPSS18.0 software (SPSS, Chicago, IL). A P value of less than 0.05 was considered significant.
Results
The Expression of RP11-166P13.3 and LINC00511 Were Upregulated in ER Positive Ovarian Cancer Cell Lines
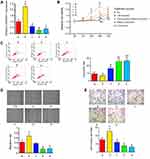
By using ChIPBase (http://rna.sysu.edu.cn/chipbase/), bioinformatics analysis showed that ESR1 is involved in the regulation of 175 lncRNAs. GEPIA is a website collecting all data from TCGA database; thus, this web is commonly used to find genes that express differently between tumor and corresponding normal tissues. GEPIA showed that many lncRNAs among the 175 lncRNAs have a significant difference in their expression between OV and normal ovarian tissues. The top 5 differential lncRNAs are MCF2L-AS1, RP11-10C24.1, RP11-166P13.3, RP4-550H1.7 and LINC00511 (Figure 1A).
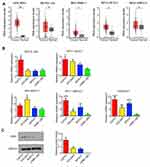 |
Figure 1 Expression of lncRNAs in OV cancer and cell lines. (A) By using ChIPBase (http://rna.sysu.edu.cn/chipbase/), bioinformatics analysis showed that ESR1 is involved in the regulation of 175 lncRNAs. GEPIA showed that many lncRNAs among the 175 lncRNAs have the significant difference in their expression between OV and normal ovarian tissues. The top 5 differential lncRNAs are MCF2L-AS1, RP11-10C24.1, RP11-166P13.3, RP4-550H1.7 and LINC00511. *P<0.05 and **P<0.01 vs normal tissues. (B) RT-qPCR assay was performed to measure the expression of MCF2L-AS1, RP11-10C24.1, RP11-166P13.3, RP4-550H1.7 and LINC00511 in OV CAOV3, OVCAR3, SKOV3 and UWB1.289 cell lines. *P<0.05, **P<0.01, ***P<0.001 vs UWB1.289 cell. (C) Western blot assay was performed to measure ESR1 expression in OV CAOV3, OVCAR3, SKOV3 and UWB1.289 cell lines. |
To determine the correlation between their expression and ERs, we measured their expression in CAOV3, OVCAR3, SKOV3 and UWB1.289 cell lines. ESR1 was expressed in the first three cell lines, but not in UWB1.289 cells23. LINC00511 and RP11-166P13.3 were up-regulated in CAOV3 (both p < 0.001), OVCAR3 (p < 0.01 and p < 0.05, respectively) and SKOV3 (p < 0.001 and p < 0.01, respectively) cells compared to those in UWB1.289 cells (Figure 1B). MCF2L-AS1 and RP11-10C24.1 were only up-regulated in CAOV3 cells compared to those in UWB1.289 cells (both p < 0.05). RP4-550H1.7 was only increased in OVCAR3 cells compared to those in UWB1.289 cells (p < 0.05). We examined the expression of ESR1 in these four types of OV cells using Western blot assay. ESR1 was expressed in CAOV3, OVCAR3 and SKOV3 cells, but not in UWB1.289 cells (Figure 1C). CAOV3 cells showed the highest ESR1 expression among these three ESR1-positive OV cells.
LINC00511 and RP11-166P13.3 Promoted the Proliferation and Invasion but Suppressed the Apoptosis of CAOV3 Cells
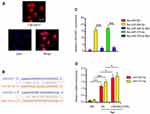
This study constructed three LINC00511-targeting shRNAs and three RP11-166P13.3-targeting shRNAs to knock down their expression in CAOV3 cells. PCR analysis showed that LINC00511-shRNA1 and RP11-166P13.3-shRNA2 caused the most significant down-regulation of LINC00511 and RP11-166P13.3 (p < 0.05, Figure 2A). Conversely, LINC00511 (p < 0.05, Figure 2B) and RP11-166P13.3 (p < 0.01) expression were increased after transfection of the expression vectors. As indicated by MTT assay, CAOV3 cell viability was dramatically decreased 48 h after transfection with LINC00511-shRNA1 (p < 0.01) and RP11-166P13.3-shRNA2 (p < 0.05, Figure 2C). CAOV3 cell viability was also reduced 72 h after transfection with LINC00511-shRNA1 (p < 0.05). After transfection of LINC00511 expression vector, CAOV3 cell viability was increased (p < 0.05 at 48 h, p < 0.01 at 72 h) compared to control cells. Increased CAOV3 cell viability was also observed 72 h after transfection with RP11-166P13.3 expression vector (p < 0.05). Silencing LINC00511 and RP11-166P13.3 increased the apoptosis rate of CAOV3 cells (p < 0.05, Figure 2D), while LINC00511 and RP11-166P13.3 overexpression had no significant effect on the apoptosis rate. LINC00511 and RP11-166P13.3 knockdown inhibited the invasion of CAOV3 cells (p < 0.05, Figure 2E), whereas LINC00511 overexpression enhanced the invasion (p < 0.05). It should be noted that LINC00511 knockdown exerted a more remarkable influence of the cell viability, apoptosis and invasion than RP11-166P13.3 knockdown. We further analyzed the correlation between LINC00511 expression and prognosis of a patient with OV using a bioinformatics analysis website (R2 platform: http://r2.amc.nl). Results showed that lower LINC00511 expression was associated to higher event-free survival probability (Figure 2F). Moreover, we analyzed the expression of LINC00511 in each stage of OV using data in GEO-GSE17260 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17260). The data showed that LINC00511 expression was remarkably higher in stage ⅢC (p < 0.01) and stage Ⅳ (p < 0.001) than in stageⅡ (Figure 2G). Based on all these data, LINC00511 was chosen for further study.
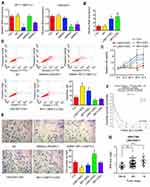 |
Figure 2 The effect of LINC00511 and RP11-166P13.3 on viability, apoptosis and invasion of CAOV3 cells. (A) CAOV-3 cells were transfected with negative control, siRNA-LINC00511 or siRNA-RP11-166P13.3. siRNA2-LINC00511 and siRNA1-RP11-166P13.3 caused the most remark reduction of LINC00511 and RP11-166P13.3 expression, respectively. PCR was performed to detect the expression of LINC00511 and RP11-166P13.3 in cells. (B) CAOV-3 cells were transfected with negative control, pEGFP-C1-LINC00511 and pEGFP-C1-RP11-166P13.3 vectors, followed by PCR measurement of LINC00511 and RP11-166P13.3 expression. After the transfection, cell viability (C), apoptosis (D) and invasion (E) were tested. LINC00511 and RP11-166P13.3 promoted CAOV3 proliferation and invasion, while inhibited apoptosis. (F) This study analyzed the correlation between LINC00511 expression and prognosis of a patient with OV using a bioinformatics analysis website (R2 platform: http://r2.amc.nl). Results showed that lower LINC00511 expression was associated to a higher event-free survival probability. (G) Moreover, the expression of LINC00511 in each stage of OV was analyzed using data in GEO-GSE17260 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17260). The data showed that LINC00511 expression was remarkably higher in stage ⅢC and stage Ⅳthan in stage Ⅱ. Data were represented as the mean ± SEM of three independent experiments. In Figure 2A–E, *P< 0.05 and **P<0.01 vs control; In Figure 2G, **P<0.01 and ***P<0.001 vs Stage Ⅱ. |
ESR1 Modulated the Expression of LINC00511 in CAOV3 Cells
According to GEPIA database, high expression of ESR1 leads to shorter overall survival (OS) (Figure 3A). Moreover, we performed univariate and multivariate analysis of clinicopathological characteristics and ESR1 expression for progression-free survival using data in GEO-GSE17260 dataset. These data suggested that ESR1 expression alone or in combination with other clinicopathological characteristics, such as tumor stages and tumor grades are prognostic predictors of OV patient survival (Table 2). Although the tumor-promoting role of ESR1 has been demonstrated, the underlying mechanism is not completely understood. By using Starbase (http://starbase.sysu.edu.cn/index.php), bioinformatics analysis showed a positive relation can be seen between ESR1 and LINC00511 (Figure 3B). The present study added the 17β-E2 and Fulvestrant to CAOV3 cells to determine the regulatory effect of ESR1 on LINC00511. Treatment with 17β-E2 to stimulate ESR1 increased LINC00511 expression (p < 0.01, Figure 3C), while ESR1 inhibitor Fulvestrant decreased LINC00511 expression (p < 0.05). As indicated by ChIP assay, the treatment with 17β-E2 increased the amount of ESR1 that bound to the promoter region of LINC00511 (p < 0.05, Figure 3D). Conversely, treatment with Fulvestrant reduced the amount of ESR1 that bound to the promoter region of LINC00511 (p < 0.05).
 |
Table 2 Univariate and Multivariate Analysis of Clinicopathological Characteristics and ESR1 Expression for Progression-Free Survival |
The Regulatory Effects of ESR1 on Growth Hallmarks of CAOV3 Cells Was Dependant on LINC00511
To determine that the regulatory effects of ESR1 on growth hallmarks of CAOV3 cells were associated to LINC00511, we treated CAOV3 cells with 17β-E2 and knocked LINC00511 down in combination. PCR analysis showed that silencing LINC00511 reversed the increase of LINC00511 induced by 17β-E2 (p < 0.05, Figure 4A). Treatment with 17β-E2 for 48 h increased the CAOV3 cell viability (p < 0.05, Figure 4B), while silencing LINC00511 conversely decreased the cell viability after 17β-E2 treatment. LINC00511 knockdown and Fulvestrant treatment alone decreased CAOV3 cell viability (p < 0.05). Apoptosis rate of CAOV3 cells was decreased after 17β-E2 treatment (p < 0.05, Figure 4C), but increased after LINC00511 knockdown and Fulvestrant treatment alone (p < 0.05). LINC00511 knockdown not only blocked the decrease of apoptosis rate induced by 17β-E2, but also increased the apoptosis. Treatment with 17β-E2 enhanced the migration and invasion of CAOV3 cells (p < 0.05, Figure 4D and E); however, LINC00511 knockdown disrupted the promoting effect of 17β-E2. Conversely, both LINC00511 knockdown and Fulvestrant treatment inhibited the migration and invasion of CAOV3 cells (p < 0.05).
This study also evaluated the effect of LINC00511 on the growth of the transplant CAOV3 tumor in nude mice. Silencing LINC00511 inhibited the growth of CAOV3 tumor in nude mice (p < 0.05, Figure 5A). Moreover, the Ki67 staining in the CAOV3 tumor tissues was also attenuated after the knockdown of LINC00511 (Figure 5B).
LINC00511 Inhibited Expression of Several miRNAs in CAOV3 Cells
FISH assay demonstrated that LINC00511 was located both in the nucleus and cytoplasm (Figure 6A). Starbase 3.0 analysis showed the interaction of LINC00511 with miR-424-5p and miR-370-5p (Figure 6B). The interaction was confirmed by RNA-pull down assay, in which miR-424-5p and miR-370-5p probes pulled down LINC00511; however, miR-424-5p and miR-370-5p Mut probes failed to pull down LINC00511 (Figure 6C). As indicated RIP assay, miR-424-5p and miR-370-5p were able to interact with Ago protein. Knockdown of LINC00511 enhanced the interaction of miR-424-5p and miR-370-5p with Ago protein (p < 0.05, Figure 6D).
Discussion
ESR1 is an important component of the estrogen receptor. Estrogen receptor can function as a ligand-activated transcription factor regulating gene expression. It has been extensively reported of the pathogenic role of ESR1 especially in hormone-dependent cancers such as breast cancer,14 endometrial cancer15 as well as OV cancer.16 However, the underlying pathogenic mechanism is not completely understood. It is generally believed that ESR1 regulates the expression of protein-coding genes that are closely related to cell proliferation, survival and differentiation, therefore participates in cancer processes.14,15 However, this study found that ESR1 is also involved in the transcript regulation of a class of non-coding genes, these genes can produce lncRNAs. The molecular functions of LncRNAs are varied, including acting as scaffolds, decoys, signal transducers, and regulators in cis or trans.11–13 These functions render LncRNAs regulate gene expression in multiple manners. In this study, ESR1 was involved in the regulation of some lncRNAs, which suggested the complex regulatory functions of ESR1 in cancer and other diseases.
This study found that two LncRNAs, LINC00511 and RP11-166P13.3, were implicated in the regulation of OV growth properties. Knockdown of both LINC00511 and RP11-166P13.3 inhibited the viability and invasion of CAOV3 cells but increased the apoptosis rate. Of note, LINC00511 knockdown exerted a more remarkable influence of the cell viability, apoptosis and invasion than RP11-166P13.3 knockdown. Previous studies reported that LINC00511 is upregulated in breast cancer, non-small-cell lung cancer, squamous cell carcinoma and pancreatic ductal adenocarcinoma.16–19 In these types of cancers, LINC00511 acts as an oncogene affecting tumorigenesis, tumor size, and metastasis. In non-small-cell lung cancer, the cancer-promoting effect of LINC00511 is associated to the suppression of p57 expression by regulating histone methyltransferase EZH2.20
We found that LINC00511 was expressed in ESR1-expressing OV cells but not in OV cells lost of ESR1 expression. Moreover, LINC00511 showed the highest expression in CAOV3 cells among three ESR1-expressing OV cells (CAOV3, OVCAR3 and SKOV3). CAOV3 cells also showed the higher ESR1 expression than OVCAR3 and SKOV3 cells. These results implied a positive correlation between ESR1 and LINC00511 expression. Bioinformatics analysis and following ChIP assay revealed that ESR1 was able to bind to the promoter region of LINC00511 gene. To determine the regulatory effect of ESR1 on LINC00511, we treated CAOV3 cells with ESR1 activator and inhibitor. Treatment with 17β-E2 increased LINC00511 expression, while Fulvestrant decreased LINC00511 expression. Transfection of siRNA-LINC00511 not only blocked the increase of LINC00511 expression caused by 17β-E2, but also abrogated promoting the effect of 17β-E2 on OV cell viability and invasion. These results suggested that LINC00511 plays an important role in ESR1 procarcinogenic effects on OV.
This study further investigated the mechanism underlying the cancer-promoting effects of LINC00511. As indicated by FISH assay, LINC00511 was located in the cytoplasm and nucleus of CAOV3 cells. LncRNAs in the cytoplasm is able to function as competing endogenous RNAs (ceRNAs) of miRNA to interfere with miRNA-mediated degradation of mRNA. Bioinformatics analysis revealed the interaction of LINC00511 with miR-424-5p and miR-370-5p, which was identified by RNA-pull down assay. As indicated by RIP assay, silencing LINC00511 increased the interaction of LINC00511 with miR-424-5p and miR-370-5p. It has been identified that miR-424-5p and miR-370-5p can target various mRNA of oncogenes, thus exerting anti-cancer effects in OV.21–23 Therefore, ESR1-up-regulated LINC00511 probably increases oncogene expression by disrupting miR-424-5p and miR-370-5p.
In conclusion, this study found that ESR1 transcriptionally regulated LINC00511 expression. Up-regulated LINC00511 promoted the viability, migration and invasion of CAOV3 cells with the inhibition of the apoptosis. These cancer-promoting effects are probably associated to the interruption of miR-424-5p and miR-370-5p. This study added further understanding of procarcinogenic role of ESR1 in OV.
Acknowledgments
This work is supported by Hainan key Program of Research and Development (ZDYF2017088).
Author Contributions
Kang Wang, Genhai Zhu and Shan Bao performed the experiments. Kang Wang, Genhai Zhu, Shan Bao and Shiling Chen contributed to conception and design, acquisition of data, or analysis and interpretation of data; Kang Wang, Genhai Zhu, Shan Bao and Shiling Chen drafted the article. All author approved the version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Romero I, Bast RC
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.v69.1
3. Modugno F, Laskey R, Smith AL, Andersen CL, Haluska P, Oesterreich S. Hormone response in ovarian cancer: time to reconsider as a clinical target? Endocr Relat Cancer. 2012;19(6):R255–279. doi:10.1530/ERC-12-0175
4. Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson JA. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci U S A. 1996;93(12):5925–5930. doi:10.1073/pnas.93.12.5925
5. Mosselman S, Polman J, Dijkema R. ER beta: identification and characterization of a novel human estrogen receptor. FEBS Lett. 1996;392(1):49–53. doi:10.1016/0014-5793(96)00782-X
6. Li S, Li Y, Wen Z, Kong F, Guan X, Liu W. microRNA-206 overexpression inhibits cellular proliferation and invasion of estrogen receptor alpha-positive ovarian cancer cells. Mol Med Rep. 2014;9(5):1703–1708. doi:10.3892/mmr.2014.2021
7. Wang S, Li X, Zhang W, et al. Genome-wide investigation of genes regulated by ERalpha in breast cancer cells. Molecules. 2018;23:10.
8. Wang Y, Lu Y, Li Z, et al. Oestrogen receptor alpha regulates the odonto/osteogenic differentiation of stem cells from apical papilla via ERK and JNK MAPK pathways. Cell Prolif. 2018;51(6):e12485. doi:10.1111/cpr.12485
9. Peng Z, Zhang C, Duan C. Functions and mechanisms of long noncoding RNAs in lung cancer. Onco Targets Ther. 2016;9:4411–4424. doi:10.2147/OTT.S109549
10. Zuo Y, Li Y, Zhou Z, Ma M, Fu K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed Pharmacother. 2017;95:922–928. doi:10.1016/j.biopha.2017.09.005
11. Wu J, Zhao W, Wang Z, Xiang X, Zhang S, Liu L. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J Cell Mol Med. 2019;23(1):680–688. doi:10.1111/jcmm.2019.23.issue-1
12. Zou MF, Ling J, Wu QY, Zhang CX. Long non-coding RNA PVT1 functions as an oncogene in ovarian cancer via upregulating SOX2. Eur Rev Med Pharmacol Sci. 2018;22(21):7183–7188. doi:10.26355/eurrev_201811_16251
13. Liu X, Wen J, Wang H, Wang Y. Long non-coding RNA LINC00460 promotes epithelial ovarian cancer progression by regulating microRNA-338-3p. Biomed Pharmacother. 2018;108:1022–1028. doi:10.1016/j.biopha.2018.09.103
14. Hu HB, Chen Q, Ding SQ. LncRNA LINC01116 competes with miR-145 for the regulation of ESR1 expression in breast cancer. Eur Rev Med Pharmacol Sci. 2018;22(7):1987–1993. doi:10.26355/eurrev_201804_14726
15. Toderow V, Rahmeh M, Hofmann S, et al. Promotor analysis of ESR1 in endometrial cancer cell lines, endometrial and endometriotic tissue. Arch Gynecology Obstetrics. 2017;296(2):1–8. doi:10.1007/s00404-017-4405-x
16. Czogalla B, Kahaly M, Mayr D, et al. Interaction of ERα and NRF2 impacts survival in ovarian cancer patients. Int J Mol Sci. 2018;20(1):E112. doi:10.3390/ijms20010112
17. Lu G, Li Y, Ma Y, et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res. 2018;37(1):289. doi:10.1186/s13046-018-0945-6
18. Ding J, Yang C, Yang S. LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2. J Oral Pathol Med. 2018;47(5):468–476. doi:10.1111/jop.2018.47.issue-5
19. Zhao X, Liu Y, Li Z, et al. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2018;22(1):655–667. doi:10.1111/jcmm.2018.22.issue-1
20. Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol Ther Nucleic Acids. 2016;5(11):e385. doi:10.1038/mtna.2016.94
21. Hua F, Li CH, Chen XG, Liu XP. Long noncoding RNA CCAT2 knockdown suppresses tumorous progression by sponging miR-424 in epithelial ovarian cancer. Oncol Res. 2018;26(2):241–247. doi:10.3727/096504017X14953948675412
22. Wu X, Ruan Y, Jiang H, Xu C. MicroRNA-424 inhibits cell migration, invasion, and epithelial mesenchymal transition by downregulating doublecortin-like kinase 1 in ovarian clear cell carcinoma. Int J Biochem Cell Biol. 2017;85:66–74. doi:10.1016/j.biocel.2017.01.020
23. Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids. 2018;8:22.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.