Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA LINC00239 Functions as a Competitive Endogenous RNA by Sponging microRNA-484 and Enhancing KLF12 Expression to Promote the Oncogenicity of Colorectal Cancer
Authors Luo X, Yue M, Li C, Sun D, Wang L
Received 24 August 2020
Accepted for publication 21 October 2020
Published 23 November 2020 Volume 2020:13 Pages 12067—12081
DOI https://doi.org/10.2147/OTT.S278582
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tohru Yamada
This paper has been retracted.
Xiaofan Luo, Meng Yue, Chenguang Li, Di Sun, Lei Wang
Department of Colorectal and Anal Surgery, First Hospital of Jilin University, Changchun, Jilin 130021, People’s Republic of China
Correspondence: Lei Wang
Department of Colorectal and Anal Surgery, First Hospital of Jilin University, 71 Xinmin Road, Changchun, Jilin 130021, People’s Republic of China
Email [email protected]
Background: Long intergenic non-protein coding RNA 239 (LINC00239) is an oncogenic long non-coding RNA in acute myeloid leukemia. We aimed to determine LINC00239 expression in colorectal cancer (CRC) and examine the influences of LINC00239 on tumor behaviors of CRC cells. Furthermore, the mechanism underlying the actions of LINC00239 in CRC was unveiled in detail.
Materials and Methods: Quantitative real-time polymerase chain reaction was used to detect LINC00239 expression in CRC tissues and cell lines. CRC cell proliferation, apoptosis, migration, and invasion were investigated by cell counting kit-8 assays, flow cytometry, and cell migration and invasion assays, respectively. Tumor xenograft experiments were performed to evaluate the tumor growth of CRC cells in vivo. The interactions among LINC00239, microRNA-484 (miR-484), and kruppel-like factor 12 (KLF12) were analyzed by bioinformatics prediction, RNA immunoprecipitation and luciferase reporter assay.
Results: LINC00239 was upregulated in CRC tissues and cell lines. LINC00239 knockdown impaired CRC cell proliferation, migration, and invasion and promoted apoptosis in vitro. Additionally, LINC00239 deficiency inhibited CRC growth in vivo. Mechanistically, LINC00239 functioned as a competing endogenous RNA by directly sponging miR-484, thereby enhancing KLF12 expression. Rescue experiments further corroborated that miR-484 inhibition or KLF12 overexpression reversed the inhibitory actions of LINC00239 knockdown in CRC cells.
Conclusion: The LINC00239/miR-484/KLF12 pathway executed critical roles in CRC oncogenicity and may provide potential targets for CRC treatments.
Keywords: long intergenic non-protein coding RNA 239, kruppel-like factor 12, CRC, miRNA sponge
Expression of Concern for this paper has been published
Introduction
Colorectal cancer (CRC) is the third most common human cancer and the second leading cause of cancer-related mortalities worldwide,1 contributing to ~1.2 million new cases and 860,000 deaths each year.2 In recent years, the incidence and mortality of CRC have continuously increased in China.3 Although great efforts have been made to identify effective diagnostic methods and anticancer treatments, the outcomes of CRC patients remain unsatisfactory, and a significant proportion of patients still die of CRC.4,5 About 50% of patients with CRC present with metastasis at their initial diagnosis or recurrence and metastasis after treatment with first-line therapies.6 Colorectal carcinogenesis and progression are complex and multifactorial processes that involve gene mutations, chronic inflammation, colorectal adenoma, lifestyle factors, and genetics.7,8 As a result, the detailed mechanisms responsible for CRC pathogenesis are largely unclear and remain to be elucidated. Hence, it is particularly urgent to comprehensively investigate the molecular events underlying CRC progression to develop promising approaches for the management of CRC.
About 98% of the human transcriptome is composed of non-coding RNAs,9,10 suggesting that these RNAs may play a significant role in diverse pathophysiological processes. Long non-coding RNAs (lncRNAs) are a class of non-coding transcripts longer than 200 nucleotides.11 They lack protein-coding capacity but perform important functions in chromatin modification, transcriptional regulation, and post-transcriptional regulation.12 Additionally, lncRNAs execute crucial actions in almost all types of cancer-related processes. Specifically, several lncRNAs are aberrantly expressed in CRC and play vital roles in cancer genesis and progression by controlling various biological processes.13–15
MicroRNAs (miRNAs) are another group of non-coding and short RNA molecules with approximately 17–25 nucleotides.16 MiRNAs negatively regulate gene expression by triggering translation suppression and/or mRNA degradation via binding to the 3ʹ-untranslated regions (3ʹ-UTRs) of their target genes.17 MiRNAs play cancer-inhibiting or cancer-promoting actions to regulate the oncogenicity of CRC and are implicated in the modulation of multiple tumor cell behaviors.18,19 LncRNAs have miRNA binding sites and can function as competing endogenous RNAs (ceRNAs) by sponging miRNAs, thus modulating the expression of miRNA targets.20 Therefore, an in-depth investigation of cancer-associated lncRNAs and miRNAs in CRC may provide useful information for identifying potential targets for cancer diagnosis and therapy.
Long intergenic non-protein coding RNA 239 (LINC00239) was previously demonstrated as an oncogenic lncRNA in acute myeloid leukemia.21 However, the exact roles of LINC00239 in CRC and the mechanism by which it regulates CRC progression have not yet been studied. To address this, our current study aimed to determine the expression profile of LINC00239 in CRC and examined the roles of LINC00239 in regulating CRC cell behavior. Furthermore, the mechanism underlying the actions of LINC00239 in CRC was explored in detail.
Materials and Methods
Patients and Clinical Tissues
Sixty-three pairs of CRC tissues and matched adjacent normal tissues were obtained from patients in the First Hospital of Jilin University. All patients had not been diagnosed with other cancer types and did not receive chemotherapy or radiotherapy prior to surgery. A total of 17 patients with CRC were diagnosed at stage III/IV. We informed the advantage of neoadjuvant therapy to these patients. They insisted for surgical resection, and all these patients received adjuvant therapy after surgery. All tissues were immersed in liquid nitrogen immediately after surgical excision and stored in liquid nitrogen until use. The Ethics Committee of the First Hospital of Jilin University approved this study (FHJLU.2018–0614), and the experimental steps were conducted in accordance with the Declaration of Helsinki. Written informed consent was also obtained from all participants.
Cell Lines
A normal human colon epithelium cell line (FHC; RRID:CVCL_3688) was purchased from the American Type Culture Collection (Manassas, VA, USA) and grown in DMEM:F12 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific Inc.), 25 mM HEPES, 10 ng/mL cholera toxin, 0.005 mg/mL insulin, 0.005 mg/mL transferrin, 100 ng/mL hydrocortisone, and 20 ng/mL human recombinant epidermal growth factor.
All five human CRC cell lines, including SW480 (RRID:CVCL_0546), SW620 (RRID:CVCL_0547), DLD-1 (RRID:CVCL_0248), HCT116 (RRID:CVCL_0291), and HT-29 (RRID:CVCL_0320), were obtained from the Shanghai Institute for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). SW480 and SW620 cells were maintained in L-15 medium (Gibco; Thermo Fisher Scientific Inc.) supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific Inc.). McCoy’s 5A medium (Gibco; Thermo Fisher Scientific Inc.) was used to culture HCT116 and HT29 cell lines, and DLD-1 cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific Inc.). Both media were supplemented with 10% FBS and 1% penicillin/streptomycin. All cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
Cell Transfection
Three small interfering RNAs (siRNAs) targeting LINC00239 (si-LINC00239#1, #2, and #3) and negative control (NC) siRNA (si-NC) were synthesized by Shanghai GenePharma Inc. (Shanghai, China). MiR-484 mimic, NC mimic, miR-484 inhibitor, and NC inhibitor were all purchased from Guangzhou RiboBio Inc. (Guangzhou, China). The KLF12 overexpression vector pcDNA3.1-KLF12, LINC00239 overexpression plasmid pcDNA3.1-LINC00239 and empty pcDNA3.1 vector were bought from GeneChem Co., Ltd. (Shanghai, China). CRC cells were seeded into 6-well plates, grown to 70%–80% confluence, and transfected with the abovementioned oligonucleotides or vectors using Lipofectamine® 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific).
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
TRIzol Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used for total RNA extraction. The quality and concentration of total RNA were examined by measuring the absorbance at 260 and 280 nm using a Nanodrop Spectrophotometer (Invitrogen; Thermo Fisher Scientific, Inc.). The one Step TB Green® PrimeScript™ PLUS RT-PCR Kit (Takara Biotechnology CO., LTD., Dalian, China) was applied to determine the expression of miR-484, with U6 small nuclear as a reference.
To quantify LINC00239 and KLF12 expression, reverse transcription was conducted using a PrimeScript™ RT reagent Kit with gDNA Eraser (Takara Biotechnology Co., Ltd.), after which quantitative PCR was performed using a TB Green® Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.). Glycerol-3-phosphate dehydrogenase (GAPDH) was used as an internal control for normalizing LINC00239 and KLF12 levels. All reactions were conducted on an Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Foster City, USA). Relative gene expression was analyzed according to the 2−ΔΔCt method.
Cell Counting Kit-8 (CCK-8) Assay
Transfected cells were seeded into 96-well plates at a density of 2,000 cells per well. Cells were incubated at 37°C with 5% CO2 for 0, 1, 2, and 3 days, and cell proliferation was tested at each designated time point. A volume of 10 µL CCK-8 reagent (Beyotime Institute of Biotechnology; Shanghai, China) was added to cells and incubated at 37°C for 2 h. The absorbance at 450 nm was read using a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA), and growth curves were plotted accordingly.
Flow Cytometry Analysis
The apoptotic rate was detected with an Annexin V-FITC Apoptosis Detection Kit (Beyotime Institute of Biotechnology). Transfected cells were harvested by trypsin digestion after 48 h of incubation, washed with ice-cooled phosphate-buffered solution, and centrifugated. Next, cells were resuspended in Annexin V-FITC binding buffer (195 μL), and then 5 μL Annexin V-FITC and 10 μL propidium iodide were added and incubated for 15 min at room temperature in the dark. The percentage of apoptotic cells was determined via a FACScan flow cytometer (BD Biosciences, San Jose, CA, USA).
Cell Migration and Invasion Assays
The cell migration assay was performed using Transwell chambers with an 8-μm polycarbonate membrane (BD Biosciences). Transfected cells were collected by trypsin digestion, centrifuged, and resuspended in FBS-free culture medium. The upper chambers were covered with a 200-μL cell suspension containing 1 × 105 cells. The lower chambers were filled with 500 μL culture medium containing 10% FBS. Twenty-four hours later, the non-migrated cells were gently removed with a cotton swab, whereas the migrated cells were fixed with paraformaldehyde and stained with 0.1% crystal violet. After extensive washing, images were taken under an inverted light microscope (Olympus, Tokyo, Japan). Six visual fields were randomly selected, and the number of migrated cells was counted. To detect cell invasion, the chambers were pre-coated with Matrigel (BD Biosciences), and the subsequent experimental steps were the same as those described in the migration assay.
Tumor Xenograft Experiments
Animal studies were conducted under the approval of the Ethics Committee of Animal Experiments of the First Hospital of Jilin University (FHJLU.2019–0201) and performed in compliance with the NIH guidelines for the care and use of laboratory animals. Short hairpin RNAs (shRNAs) targeting LINC00239 (sh-LINC00239) and NC shRNA (sh-NC) were designed and synthesized by Shanghai GenePharma Inc. and ligated into a GenePharma Supersilencing Vector (Shanghai GenePharma Inc.). The lentivirus stably expressing sh-LINC00239 or sh-NC was transfected into SW480 cells, and the stably transfected cells were selected using puromycin. BALB/c male nude mice (4–5 weeks) were purchased from the Laboratory Animal Center of Southern Medical University (Guangzhou, China) and subcutaneously injected with SW480 cells stably expressing sh-LINC00239 or sh-NC. The volume of subcutaneous xenografts was determined weekly by measuring tumor width and length and calculated with the following formula: volume = 1/2 (length × width2). Four weeks after tumor cell injection, mice were euthanized, and tumor xenografts were obtained for weight measurements and molecular analyses.
Bioinformatics Analysis
The online public algorithm miRDB (http://mirdb.org/) was applied to identify the miRNAs that directly interact with LINC00239. The putative target genes of miR-484 were searched using three algorithms, including Targetscan (http://www.targetscan.org/vert_60/) and TargetMiner (http://www.isical.ac.in/~bioinfo_miu).
Subcellular Fractionation Assay
This assay was performed to examine the subcellular distribution of LINC00239 in CRC cells. Using a Cytoplasmic and Nuclear RNA Purification Kit (Norgen, Thorold, ON, Canada), the cytoplasm and nucleus fractions of CRC cells were separated, and both fractions were subjected to RNA extraction and qRT-PCR. GAPDH and U6 small nuclear RNA served as the cytoplasmic and nuclear controls, respectively.
RNA Immunoprecipitation (RIP) Assay
The Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) was used according to the manufacturer’s protocol. Briefly, CRC cells were lysed in RIP buffer, and the cell lysate was treated with magnetic beads conjugated with an Ago2 antibody and normal mouse IgG (Millipore). Then, the magnetic beads were harvested and rinsed with wash buffer. After removing the proteins using Proteinase K, the co-precipitated RNA was isolated, and qRT-PCR analysis was conducted to determine the enrichment of LINC00239 and miR-484.
Luciferase Reporter Assay
LINC00239 fragments containing wild-type (WT) or mutant (MUT) miR-484 binding sites were synthesized and cloned into the pmirGLO Dual-Luciferase Vector (Promega Corporation, Madison, WI, USA) to generate LINC00239-WT and LINC00239-MUT reporter vectors. Similarly, KLF12-WT and KLF12-MUT reporter vectors were obtained using the same experimental steps. For luciferase reporter assays, CRC cells were inoculated into 24-well plates and cotransfected with the WT or MUT reporter vector and miR-484 mimic or NC mimic using Lipofectamine® 2000 transfection reagent. Forty-eight hours later, luciferase activity was determined with the Dual-Luciferase Reporter Assay System (Promega).
Western Blot Analysis
Cultured cells were collected and lysed with RIPA lysis buffer (Beyotime Institute of Biotechnology). An enhanced BCA protein assay kit was used for total protein quantification. Equal amounts of total protein were separated by sodium dodecyl sulfate-polyacrylamide electrophoresis on 10% gels and transferred to polyvinylidene fluoride membranes. After 2 h of blocking with 5% nonfat powdered milk, the membranes were incubated with primary antibodies against KLF12 (1:1,000 dilution; sc-134,373; Santa Cruz Biotechnology, CA, USA) or GAPDH (1:1,000 dilution; sc-51907; Santa Cruz Biotechnology) overnight at 4°C. The membranes were further incubated for 2 h with a goat anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (1:5000 dilution; sc-516102; Santa Cruz Biotechnology) at room temperature, and the protein bands were detected using Thermo Scientific Pierce ECL Plus Substrate (Pierce; Thermo Fisher Scientific, Inc.). GAPDH served as an internal control for KLF12 expression.
Statistical Analysis
The SPSS version 20 software package (IBM SPSS Inc, Chicago, IL) was used for statistical analysis. The difference between two groups was assessed with Student’s t-test, and a one-way analysis of variance (ANOVA) followed by Tukey’s test was used to compare multiple groups. The correlation between LINC002239 and miR-484 expression was assessed by Pearson’s coefficient analysis. All results were presented as means ± standard deviations, and a value of P < 0.05 was considered statistically significant.
Results
LINC00239 Depletion Impairs CRC Cell Proliferation, Migration, and Invasion and Promotes Cell Apoptosis in vitro
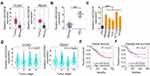
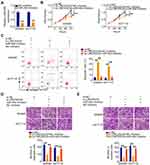
First, the expression of LINC00239 was analyzed in colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) using TCGA and GTEx databases. LINC00239 was found to be strikingly overexpressed in both COAD and READ (Figure 1A). Additionally, LINC00239 expression was measured in 63 pairs of CRC tissues and matched adjacent normal tissues. qRT-PCR analysis indicated that LINC00239 expression was higher in CRC tissues than that in adjacent normal tissues (Figure 1B). Consistently, all five CRC cell lines (SW480, SW620, DLD-1, HCT116, and HT-29) exhibited higher expression of LINC00239 in comparison with that in FHC cells (Figure 1C). Correlation analysis showed that high LINC00239 levels were correlated with tumor stage in READ patients (Figure 1D; P = 0.0284), but no obvious relationship was identified between LINC00239 expression and tumor stage in COAD (P = 0.957). Furthermore, the analysis of TCGA and GTEx databases revealed that an increased level of LINC00239 was not associated with either overall survival (Figure 1E; P = 0.41) or disease-free survival (Figure 1F; P = 0.21) in CRC.
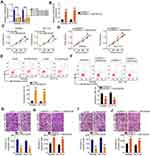
To research the roles of LINC00239 in CRC, LINC00239 expression was silenced (Figure 2A) or overexpressed (Figure 2B) in SW480 and HCT116 cells using si-LINC00239 or pcDNA3.1-LINC00239, respectively. The analysis of silencing efficiency showed that si-LINC00239#1 presented the highest inhibitory effect on LINC00239 expression in SW480 and HCT116 cells; thus, si-LINC00239#1 was selected for follow-up experiments. The effect of LINC00239 silencing or overexpression on CRC cell proliferation was examined by the CCK-8 assay. Loss of LINC00239 obviously hindered the proliferation of SW480 and HCT116 cells (Figure 2C), while LINC00239 upregulation increased cell proliferation (Figure 2D). In addition, the apoptosis rate of LINC00239-deficient SW480 and HCT116 cells was clearly increased (Figure 2E), as evidenced by flow cytometry analysis. In contrast, transfection with pcDNA3.1-LINC00239 significantly decreased cell apoptosis (Figure 2F). Furthermore, the downregulation of LINC00239 impaired the migratory (Figure 2G) capacity of SW480 and HCT116 cells compared with si-NC-transfected cells, whereas upregulation of LINC00239 showed the opposite effect (Figure 2H). Moreover, transfection with si-LINC00239 harmed the invasive ability of SW480 and HCT116 cells (Figure 2I), whereas transfection with pcDNA3.1-LINC00239 promoted cell invasion (Figure 2J). Thus, these results demonstrated that LINC00239 exerted a pro-cancer role in CRC oncogenesis and progression.
LINC00239 Serves as an Endogenous Molecular Sponge for miR-484 in CRC Cells
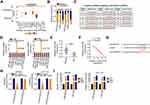
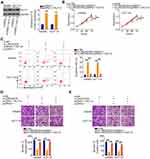
To elucidate the mechanisms by which LINC00239 affects CRC progression, lncATLAS (http://lncatlas.crg.eu/) was used to assess the localization of LINC00239. LINC00239 localization was predicted to be mainly cytoplasmic (Figure 3A), which was further verified by subcellular fractionation assays (Figure 3B). Accumulating studies have uncovered that cytoplasmic lncRNAs can directly bind to miRNA and act as a miRNA sponge.22–24 Following bioinformatics prediction, 23 miRNAs were identified to have complementary sites within LINC00239 (Figure 3C). Of these candidates, miR-629-3p,25 miR-106a-3p,26 miR-660-5p,27 miR-484,28 miR-320b/c/d,29,30 and miR-320a-3p31 were selected for follow-up studies because of their important roles in carcinogenesis and cancer progression.
qRT-PCR was conducted to explore whether these miRNAs can be modulated by LINC00239 in CRC cells. The results revealed that LINC00239 depletion increased miR-484 expression in SW480 and HCT116 cells but had no impact on the expression of the other seven miRNAs (Figure 3D). Moreover, miR-484 expression was reduced in CRC tissues (Figure 3E) and inversely correlated with LINC00239 levels (Figure 3F; r = −0.7378, P < 0.0001). The binding sites of miR-484 within LINC00239 were presented in Figure 3G. To further investigate the physical interactions between miR-484 and LINC00239, a luciferase reporter assay was conducted in SW480 and HCT116 cells cotransfected with miR-484 mimic or miR-NC and LINC00239-WT or LINC00239-MUT. As shown in Figure 3H, the upregulation of miR-484 obviously decreased LINC00239-WT luciferase activity in SW480 and HCT116 cells, whereas the luciferase activity of LINC00239-MUT was unaffected after miR-484 overexpression. Furthermore, RIP assay results confirmed that LINC00239 and miR-484 were notably enriched by Ago2 in SW480 and HCT116 cells (Figure 3I), revealing that they existed in the same RNA-induced silencing complex. Collectively, LINC00239 acted as a miR-484 sponge in CRC cells.
MiR-484 Plays Anti-Oncogenic Roles and Directly Targets KLF12 in CRC Cells
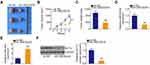
The detailed roles of miR-484 were also examined in CRC cells. After confirming the overexpression efficiency of miR-484 mimic (Figure 4A), CCK-8 assays, flow cytometry analysis, and cell migration and invasion assays were performed to detect the influences of miR-484 overexpression on CRC cells. The experimental results confirmed that ectopic miR-484 expression effectively suppressed SW480 and HCT116 cell proliferation (Figure 4B) but promoted cell apoptosis (Figure 4C). Furthermore, the migration (Figure 4D) and invasion (Figure 4E) of SW480 and HCT116 cells were apparently decreased following miR-484 mimic transfection.
To determine the detailed mechanisms underlying the actions of miR-484 in CRC cells, bioinformatics analysis was used to identify the potential targets of miR-484. A miR-484 binding site was found in the 3ʹ-UTR of KLF12 (Figure 4F), which was selected for subsequent verification because of its critical oncogenic roles in CRC.32,33 To investigate this possibility, a luciferase reporter assay was conducted, and the results revealed that miR-484 evidently reduced the luciferase activity of KLF12-WT in SW480 and HCT116 cells without influencing the activity of the LKF12-MUT reporter (Figure 4G). By measuring KLF12 expression, we revealed that KLF12 was overexpressed in CRC tissues compared with adjacent normal tissues (Figure 4H). In addition, an inverse correlation between KLF12 mRNA and miR-484 expression in CRC tissues was confirmed by Pearson’s correlation analysis (Figure 4I; r = −0.6702, P < 0.0001). Then, qRT-PCR and Western blotting analyses uncovered that the mRNA (Figure 4J) and protein (Figure 4K) levels of KLF12 were suppressed by miR-484 upregulation in SW480 and HCT116 cells. Together, these data suggested that miR-484 played cancer-inhibiting functions in CRC cells and directly targeted KLF12.
LINC00239 Sponges miR-484 and Enhances KLF12 Expression in CRC Cells
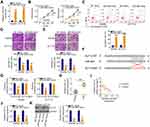
The above results showed that LINC00239 acted as a miR-484 sponge and that miR-484 directly targeted KLF12 in CRC cells. Thus, whether LINC00239 subsequently controls KLF12 expression was explored. KLF12 expression was determined in SW480 and HCT116 cells after LINC00239 knockdown. Intriguingly, both the mRNA (Figure 5A) and protein (Figure 5B) levels of KLF12 were reduced in LINC00239-deficient SW480 and HCT116 cells. RIP assay results indicated that LINC00239, miR-484, and KLF12 were all enriched by Ago2 antibody precipitation (Figure 5C), suggesting the co-existence of LINC00239, miR-484, and KLF12 in an RNA-induced silencing complex. Pearson’s correlation analysis further validated a positive correlation between LINC00239 and KLF12 mRNA expression in CRC tissues (Figure 5D; r = 0.6402, P < 0.0001). Furthermore, rescue experiments revealed that the LINC00239 silencing-mediated downregulation of KLF12 mRNA (Figure 5E) and protein (Figure 5F) was restored by miR-484 inhibition. Altogether, these findings suggested that LINC00239 increased KLF12 expression in CRC cells by sequestering miR-484.
MiR-484 Inhibition or KLF12 Overexpression Restores the Inhibitory Effects of LINC00239 Knockdown in CRC Cells
Rescue assays were conducted to evaluate whether the miR-484/KLF12 axis contributes to the biological effects of LINC00239 in CRC cells. To this end, the transfection efficiency of miR-484 inhibitor was first assessed via qRT-PCR. The expression of miR-484 was notably decreased after miR-484 inhibitor transfection (Figure 6A). Next, SW480 and HCT116 cells were cotransfected with si-LINC00239 and miR-484 inhibitor or NC inhibitor. LINC00239 depletion suppressed SW480 and HCT116 cell proliferation (Figure 6B) and promoted their apoptosis (Figure 6C), and the effects were abrogated when miR-484 was silenced. Additionally, miR-484 inhibition reversed the impacts induced by si-LINC00239 on the migration (Figure 6D) and invasion (Figure 6E) of SW480 and HCT116 cells.
Furthermore, the KLF12 overexpression plasmid pcDNA3.1-KLF12 was used to overexpress KLF12 (Figure 7A), and it was introduced into LINC00239-deficient SW480 and HCT116 cells. The CCK-8 assay results showed that the loss of LINC00239 significantly hindered the proliferative capacity of SW480 and HCT116 cells, whereas cotransfection with pcDNA3.1-KLF12 counteracted the effects (Figure 7B). Consistently, the increased cell apoptosis caused by si-LINC00239 was recovered by KLF12 overexpression (Figure 7C). Furthermore, KLF12 overexpression abolished the inhibitory actions of LINC00239 knockdown on the migration (Figure 7D) and invasion (Figure 7E) of SW480 and HCT116 cells. Taken together, LINC00239 aggravated the oncogenicity of CRC cells by regulating the miR-484/KLF12 axis.
Interference of LINC00239 Decreases Tumor Growth of CRC in vivo
To address the effect of LINC00239 on the tumor growth of CRC cells, mice xenograft models were constructed by inoculating SW480 cells stably expressing sh-LINC00239 or sh-NC into nude mice. The volumes of tumor xenografts in the sh-LINC00239 group were remarkably decreased compared with those in the sh-NC group (Figure 8A and B). Additionally, the sh-LINC00239 group presented reduced tumor weights compared with the sh-NC group (Figure 8C). According to qRT-PCR results, LINC00239 expression was evidently decreased (Figure 8D), whereas miR-484 was overexpressed (Figure 8E) in the LINC00239 stably depleted-tumor xenografts. Furthermore, the protein level of KLF12 in the sh-LINC00239 group was clearly downregulated compared with the sh-NC group (Figure 8F). Therefore, LINC00239 depletion impeded CRC tumor growth in vivo.
Discussion
In the past decade, lncRNAs have been identified as key regulators of carcinogenesis and cancer progression.34,35 Increasing studies have demonstrated that several lncRNAs are differentially expressed in CRC and play critical roles in CRC oncogenesis and progression.36–38 Thus, further exploration of the functions and molecular mechanisms of lncRNAs involved in regulating CRC oncogenicity is warranted and may contribute to the development of CRC therapies. Although various lncRNAs have been identified,39 only a small number of lncRNAs have been thoroughly explored in CRC. To address this important gap, our study measured the expression of LINC00239 in CRC and examined the roles of LINC00239 in regulating the oncogenicity of CRC cells in vitro and in vivo. More importantly, the detailed mechanisms responsible for the tumor-promoting activities of LINC00239 in CRC cells were elucidated.
LINC00239 is highly expressed in acute myeloid leukemia and plays oncogenic roles by promoting cell proliferation, colony-forming, and migration abilities.21 Furthermore, the upregulation of LINC00239 improves the chemoresistance of acute myeloid leukemia cells to doxorubicin.21 Additionally, LINC00239 is identified as a biomarker to predict the overall survival of patients with hepatocellular carcinoma without fibrosis.40 However, the expression and functions of LINC00239 in CRC are poorly understood. In this study, LINC00239 was found to be overexpressed in CRC tissues and cell lines. Functionally, the downregulation of LINC00239 by RNA interference restrained CRC cell proliferation, migration, and invasion and promoted cell apoptosis in vitro. Additionally, LINC00239 depletion inhibited CRC growth in vivo. These observations suggest that LINC00239 functions as an oncogene in CRC cells, providing a potential target for CRC management.
Extensive studies reported that cytoplasmic lncRNAs are important elements of the ceRNA network via their competitive binding to miRNAs and consequent protection of miRNA targets from degradation or translational inhibition.41 The molecular events of LINC00239 in promoting the aggressiveness of CRC cells remain unclear. LncATLAS, a visualization tool to obtain useful information on the localization of lncRNAs, was used to determine the subcellular distribution of LINC00239, which was further verified by subcellular fractionation assays. Our results validated for the first time that LINC000239 is a cytoplasmic lncRNA in CRC cells, suggesting that LINC00239 acts as a ceRNA or miRNA sponge. Eight miRNAs were predicted as candidates, and further analyses revealed that LINC00239 attenuated the expression of miR-484 in CRC cells. In addition, LINC00239 and miR-484 levels were inversely correlated in CRC tissues. Further, RIP and luciferase reporter assay results collectively demonstrated that LINC00239 and miR-484 exhibited a specific target-controlled relationship in CRC cells. Collectively, these results demonstrated that LINC00239 worked as a molecular sponge of miR-484 in CRC cells.
MiRNAs are known to modulate target mRNAs by directly binding to the 3ʹ-UTR of their target gene.42 Interestingly, using a combination of multiple biochemical analyses and mechanistic studies, KLF12 was identified as a direct target of miR-484 in CRC cells. Through our in-depth analysis, the relationship among LINC00239, miR-484, and KLF12 in CRC was clarified. The results indicated that LINC00239 deficiency reduced KLF12 mRNA and protein levels in CRC cells, and these regulatory actions were reversed by miR-484 inhibition. Additionally, a positive correlation between LINC00239 and KLF12 levels was observed in CRC tissues. Importantly, LINC00239, miR-484, and KLF12 were confirmed to co-exist in the same RNA-induced silencing complex. In short, these observations provided convincing evidence of a novel ceRNA pathway involving LINC00239, miR-484, and KLF12 in CRC cells.
MiR-484 is downregulated in many human cancers, including CRC,43 but its detailed roles in CRC remain elusive. Our research investigated the roles of miR-484 in regulating the malignant characteristics of CRC cells and confirmed that miR-484 is an anti-cancer miRNA. The transcription factor KLF12 was directly targeted and negatively regulated by miR-484 in CRC cells. KLF12 is known to be upregulated in CRC, and patients with high KLF12 levels show a worse prognosis compared with patients exhibiting low KLF12 expression.32 KLF12 displays tumor-promoting activities in CRC cells and participates in the regulation of multiple tumor cell behaviors.32,33 In this study, further exploration suggested that KLF12 was positively regulated by LINC00239 in CRC cells, and rescue experiments further confirmed the function of LINC00239 in sponging miR-484 to regulate KLF12 expression. Significantly, miR-484 inhibition or KLF12 overexpression effectively reversed the suppressive influences of LINC00239 silencing on the malignant phenotype of CRC cells. Therefore, our study unveiled the contribution of the LINC00239/miR-484/KLF12 pathway in regulating CRC progression.
Conclusion
To conclude, LINC00239 was upregulated in CRC and promoted the development of CRC. Mechanistically, LINC00239 sponged miR-484, which subsequently upregulated KLF12 expression in CRC cells. Our results may provide a novel method for CRC diagnosis, prevention, and anticancer therapy.
Funding
This study was not funded by any commercial or not-for-profit agencies.
Disclosure
The authors declare that they have no competing interests.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
2. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289.
3. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi:10.1136/gutjnl-2015-310912
4. Goldstein DA, Zeichner SB, Bartnik CM, Neustadter E, Flowers CR. Metastatic colorectal cancer: a systematic review of the value of current therapies. Clin Colorectal Cancer. 2016;15(1):1–6. doi:10.1016/j.clcc.2015.10.002
5. Dekker E, Rex DK. Advances in CRC prevention: screening and surveillance. Gastroenterology. 2018;154(7):1970–1984. doi:10.1053/j.gastro.2018.01.069
6. Chen Y, Xie H, Gao Q, et al. Colon cancer associated transcripts in human cancers. Biomed Pharmacother/Biomedicine & Pharmacotherapie. 2017;94:531–540. doi:10.1016/j.biopha.2017.07.073
7. Nguyen LH, Goel A, Chung DC. Pathways of colorectal carcinogenesis. Gastroenterology. 2020;158(2):291–302. doi:10.1053/j.gastro.2019.08.059
8. Tannapfel A, Neid M, Aust D, Baretton G. The origins of colorectal carcinoma: specific nomenclature for different pathways and precursor lesions. Dtsch Arztebl Int. 2010;107(43):760–766.
9. Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi:10.1038/ng.3192
10. Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126(8):2775–2782. doi:10.1172/JCI84421
11. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi:10.1016/j.cell.2009.02.006
12. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi:10.1101/gad.1800909
13. Zheng Z, Li X, You H, Zheng X, Ruan X. LncRNA SOCS2-AS1 inhibits progression and metastasis of colorectal cancer through stabilizing SOCS2 and sponging miR-1264. Aging. 2020;12(11):10517–10526. doi:10.18632/aging.103276
14. Shang A, Wang W, Gu C, et al. Long non-coding RNA CCAT1 promotes colorectal cancer progression by regulating miR-181a-5p expression. Aging. 2020;12(9):8301–8320.
15. Li T, Wang B, Zhang L, Cui M, Sun B. Silencing of long noncoding RNA LINC00346 inhibits the tumorigenesis of colorectal cancer through targeting MicroRNA-148b. Onco Targets Ther. 2020;13:3247–3257. doi:10.2147/OTT.S242715
16. Kabekkodu SP, Shukla V, Varghese VK, et al. Cluster miRNAs and cancer: diagnostic, prognostic and therapeutic opportunities. Wiley Interdiscip Rev RNA. 2020;11(2):e1563. doi:10.1002/wrna.1563
17. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840.
18. Wang H. MicroRNAs and apoptosis in colorectal cancer. Int J Mol Sci. 2020;21:15.
19. Xiao Z, Chen S, Feng S, et al. Function and mechanisms of microRNA-20a in colorectal cancer. Exp Ther Med. 2020;19(3):1605–1616.
20. Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long Noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 2019;20:22.
21. Yang Y, Dai W, Sun Y, Zhao Z. Long noncoding RNA linc00239 promotes malignant behaviors and chemoresistance against doxorubicin partially via activation of the PI3K/Akt/mTOR pathway in acute myeloid leukaemia cells. Oncol Rep. 2019;41(4):2311–2320.
22. Yan L, Li K, Feng Z, et al. lncRNA CERS6-AS1 as ceRNA promote cell proliferation of breast cancer by sponging miR-125a-5p to upregulate BAP1 expression. Mol Carcinog. 2020;59(10):1199–1208. doi:10.1002/mc.23249
23. Zhang C, Wu S, Song R, Liu C. Long noncoding RNA NR2F1-AS1 promotes the malignancy of non-small cell lung cancer via sponging microRNA-493-5p and thereby increasing ITGB1 expression. Aging. 2020;12.
24. Han Q, Li J, Xiong J, Song Z. Long noncoding RNA LINC00514 accelerates pancreatic cancer progression by acting as a ceRNA of miR-28-5p to upregulate Rap1b expression. J Exp Clin Cancer Res. 2020;39(1):151. doi:10.1186/s13046-020-01660-5
25. Li B, Meng YQ, Li Z, et al. MiR-629-3p-induced downregulation of SFTPC promotes cell proliferation and predicts poor survival in lung adenocarcinoma. Artif Cells, Nanomed Biotechnol. 2019;47(1):3286–3296. doi:10.1080/21691401.2019.1648283
26. Guo W, Li W, Yuan L, Mei X, Hu W. MicroRNA-106a-3p induces apatinib resistance and activates Janus-Activated Kinase 2 (JAK2)/Signal Transducer and Activator of Transcription 3 (STAT3) by targeting the SOCS system in gastric cancer. Med Sci Monitor. 2019;25:10122–10128. doi:10.12659/MSM.919610
27. Qi Y, Zha W, Zhang W. Exosomal miR-660-5p promotes tumor growth and metastasis in non-small cell lung cancer. J BUON. 2019;24(2):599–607.
28. Lee D, Tang W, Dorsey TH, Ambs S. miR-484 is associated with disease recurrence and promotes migration in prostate cancer. Biosci Rep. 2020;40:5. doi:10.1042/BSR20191028
29. Lv QL, Du H, Liu YL, et al. Low expression of microRNA-320b correlates with tumorigenesis and unfavorable prognosis in glioma. Oncol Rep. 2017;38(2):959–966. doi:10.3892/or.2017.5762
30. Tadano T, Kakuta Y, Hamada S, et al. MicroRNA-320 family is downregulated in colorectal adenoma and affects tumor proliferation by targeting CDK6. World J Gastrointest Oncol. 2016;8(7):532–542. doi:10.4251/wjgo.v8.i7.532
31. Zhao W, Sun Q, Yu Z, et al. MiR-320a-3p/ELF3 axis regulates cell metastasis and invasion in non-small cell lung cancer via PI3K/Akt pathway. Gene. 2018;670:31–37.
32. Kim SH, Park YY, Cho SN, Margalit O, Wang D, DuBois RN. Kruppel-like factor 12 promotes colorectal cancer growth through early growth response Protein 1. PLoS One. 2016;11(7):e0159899. doi:10.1371/journal.pone.0159899
33. Yao H, Xia D, Li ZL, et al. MiR-382 functions as tumor suppressor and chemosensitizer in colorectal cancer. Biosci Rep. 2019;39:8. doi:10.1042/BSR20180441
34. Ogunwobi OO, Mahmood F, Akingboye A. Biomarkers in colorectal cancer: current research and future prospects. Int J Mol Sci. 2020;21:15. doi:10.3390/ijms21155311
35. Jin KT, Yao JY, Fang XL, Di H, Ma YY. Roles of lncRNAs in cancer: focusing on angiogenesis. Life Sci. 2020;252:117647. doi:10.1016/j.lfs.2020.117647
36. Martinez-Barriocanal A, Arango D, Dopeso H. PVT1 long non-coding RNA in gastrointestinal cancer. Front Oncol. 2020;10:38.
37. He Q, Long J, Yin Y, et al. Emerging roles of lncRNAs in the formation and progression of colorectal cancer. Front Oncol. 2019;9:1542. doi:10.3389/fonc.2019.01542
38. Wei L, Wang X, Lv L, Zheng Y, Zhang N, Yang M. The emerging role of noncoding RNAs in colorectal cancer chemoresistance. Cell Oncol. 2019;42(6):757–768. doi:10.1007/s13402-019-00466-8
39. Grixti JM, Ayers D. Long noncoding RNAs and their link to cancer. Non-Coding RNA Research. 2020;5(2):77–82. doi:10.1016/j.ncrna.2020.04.003
40. Ye J, Li H, Wei J, et al. Risk scoring system based on lncRNA expression for predicting survival in hepatocellular carcinoma with cirrhosis. Asian Pac j Cancer Prev. 2020;21(6):1787–1795. doi:10.31557/APJCP.2020.21.6.1787
41. Zhang XZ, Liu H, Chen SR. Mechanisms of long non-coding RNAs in cancers and their dynamic regulations. Cancers. 2020;12:5. doi:10.3390/cancers12051245
42. Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96;(Suppl):R40–44.
43. Lu X, Lu J. The significance of detection of serum miR-423-5p and miR-484 for diagnosis of colorectal cancer. Clin Lab. 2015;61(1–2):187–190. doi:10.7754/Clin.Lab.2014.140625
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.