Back to Journals » OncoTargets and Therapy » Volume 12
Long Non-Coding RNA LINC00152 Regulates Cell Proliferation, Migration And Invasion In Esophageal Squamous Cell Carcinoma Via miR-107/Rab10 Axis
Received 1 July 2019
Accepted for publication 20 September 2019
Published 17 October 2019 Volume 2019:12 Pages 8553—8567
DOI https://doi.org/10.2147/OTT.S221515
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjay Singh
Zhigang Zhou,1 Fei Huang2
1Department of Thoracic Surgery, The Third Hospital of Hebei Medical University, Shijiazhuang 050051, Hebei, People’s Republic of China; 2Department of Cardio-Thoracic Surgery, The Third Hospital of Hebei Medical University, Shijiazhuang 050051, Hebei, People’s Republic of China
Correspondence: Zhigang Zhou
Department of Thoracic Surgery, The Third Hospital of Hebei Medical University, No. 139 Ziqiang Road, Shijiazhuang 050051, Hebei, People’s Republic of China
Tel +86-311-88603000
Email [email protected]
Background: Esophageal squamous cell carcinoma (ESCC) is a common malignant tumor in East Asia. Emerging evidence indicated that long intergenic non-coding RNA 152 (LINC00152) acts as an oncogene in many types of cancers including ESCC. This study aims to identify the biological mechanisms of LINC00152 in ESCC, hinting for new therapeutic target for ESCC patients.
Methods: The levels of LINC00152, microRNA-107 (miR-107) and Ras-related protein Rab-10 (Rab10) were measured in ESCC tissues and cells using qRT-PCR. The protein level of Rab10 was measured by Western blot assay. The putative target of LINC00152 or miR-107 was searched using starBase v2.0 and TargetScan online databases, and dual-luciferase reporter assay was conducted to detect the interaction between miR-107 and LINC00152 or Rab10. The cell viability was monitored by CCK8 assay, and the abilities of migration and invasion were assessed by Transwell assay, respectively. The mice model experiments were constructed to affirm the biological role of LINC00152 in vivo.
Results: LINC00152, Rab10 was significantly upregulated, and miR-107 was strikingly down-regulated in ESCC tissues and cell lines (TE-1 and KYSE30). LINC00152 was verified as a sponge for miR-107, and Rab10 was a direct target of miR-107. LINC00152 depletion decreased cell viability and abilities of migration and invasion by regulating miR-107 in vitro and blocked xenograft tumor growth in vivo. The overexpression of miR-107 reduced cell viability and the abilities of migration and invasion by modulating Rab10. LINC00152 positively regulated Rab10 expression by sponging miR-107.
Conclusion: In this study, we found that LINC00152 modulated Rab10 to promote cell proliferation, migration and invasion in ESCC by sponging miR-107. This new regulatory network may provide a novel therapeutic target for ESCC patients.
Keywords: LINC00152, miR-107, Rab10, esophageal squamous cell carcinoma
Introduction
Esophageal squamous cell carcinoma (ESCC), associates with high mortality in East Asia, is one of the histological subtypes of esophageal cancer (EC).1 Emerging evidence revealed that early diagnosis in ESCC could elevate the survival rates of ESCC patients.2 Majority of ESCC patients are diagnosed too late to get resection surgical due to the lack of biomarkers for the early detection.3 Therefore, it is urgent to find novel therapeutic targets for early detection in ESCC.
Long non-coding RNAs (lncRNAs) are a class >200 nucleotides (nt) and have no translation function. Previous documents revealed that lncRNAs closely associated with chromatin remodeling, transcriptional regulation, and post-transcriptional modulation and further affect cancer.4 Long intergenic non-coding RNA 152 (LINC00152), an 828 bp lncRNA located on human chromosome 2p11.2, was discovered in the process of demethylation during liver cancer progression.5 Plenty of evidence indicated that LINC00152 was an oncogene in various malignancies including ESCC.6 For example, a study in gastric cancer (GC) demonstrated that LINC00152 was apparently upregulated in GC tissues and cells, and LINC00152 depletion retarded cell proliferation, migration and invasion and promoted cell apoptosis.7 Actually, another document unraveled that LINC00152 was significantly increased in ESCC tissues and cells (EC109, EC9706, TE-1, TE-3, KYSE150, YSE450 and KYSE30), LINC00152 knockdown inhibited cell proliferation and induced apoptosis via miR-153-3p/FYN axis in ESCC.8 However, the full regulatory network of LINC00152 in ESCC remains to be determined.
MicroRNAs (miRNAs), a class of small RNA about 22 nt in length, have been reported to function as message RNA (mRNA) inhibitor via blocking 3ʹUTR of mRNA or degradation pathway.4 Aberrant expression of miR-107 was found in various cancers including ESCC. For instance, Zhan et al, manifested that miR-107 was dramatically decreased in osteoarthritis tissues and cells, and overexpression of miR-107 hampered cell apoptosis and impelled autophagy in osteoarthritis chondrocytes.9 Another report in ESCC revealed that miR-107 was down-regulated in ESCC cells, and its overexpressed blocked cell proliferation, migration and invasion but promoted cell apoptosis.10 Ras-related protein Rab-10 (Rab10), a member of the RAS oncogene family, has been documented to play a crucial role in vesicular trafficking and associated with many types of cancers.11 In fact, convincing evidence showed that the miR‑378a‑3p/Rab10 axis-regulated cell proliferation, invasion and migration in ESCC cells.12 Nevertheless, the biological mechanism of miR-107 or Rab10 was not fully reported in ESCC. In this study, we mainly aimed to explore the mechanism of LINC00152 in ESCC.
Materials And Methods
Patients And Tissue Specimens
Twenty-three tumor samples and its corresponding adjacent normal tissues were collected from ESCC patients in the Third Hospital of Hebei Medical University, and then stored in −80°C refrigerator until further using. All patients or these relatives in this program were provided informed consents. The ESCC patients were divided into two groups according to the level of LINC00152:high LINC00152 group (n=14), low LINC00152 group (n=11). This study was approved by the Ethics Committee of the Third Hospital of Hebei Medical University and performed in compliance with the Declaration of Helsinki Principles, which is also in accordance with the Guidelines for Animal Welfare Standards.
Cell Culture And Transfection
The ESCC cell lines (TE-1 and KYSE30) and the normal human esophageal mucosal epithelial cell line (Het-1A) were purchased from Cell Bank of the Chinese Academy of Science (Shanghai, China). All the cells were cultivated in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Rockville, MD, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) in a 37°C, 5% CO2 incubator.
Small interfering RNA (siRNA) direct targeting LINC00152 (si-LINC00152), a scramble control (si-NC), siRNA against Rab10 (si-Rab10) and its control (si-NC), miR-107 mimic (miR-107) and mock of miR-107 (miR-NC), miR-107 inhibitor (in-miR-107) and its corresponding negative control (in-miR-NC) were brought from GenePharma (Shanghai, China). Rab10 3ʹUTR or its mutant was amplified and inserted into pcDNA vector (Hanbio, Shanghai, China), namely Rab10 or pcDNA, respectively. The transient transfection was performed using Lipo-fectamine 2000 Reagent (Invitrogen, USA) referring to the manual.
RNA Isolation And Quantitative Real-Time PCR (qRT-PCR)
Total RNA in tissues and cells were extracted using miRNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. cDNA was formed using miScript RT Kit (TaKaRa, Dalian, China). The quantitative PCR was performed using SYBR Premix Ex Taq II (TaKaRa) on ABI Prism 7700 Sequence Detection System (Thermo Fisher Scientific). The relative expression levels of LINC00152, Rab10 were normalized by glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and miR-107 was normalized by small nuclear RNA U6 and calculated with the method of 2−ΔΔCt. The primers were synthesized in Songon (Shanghai, China) and listed as follows: LINC00152: (Forward, 5ʹ-GAAGGTGTCGGCAAGATC-3ʹ and Reverse, 5ʹ-TCGGTGTCTGTCATATTCG-3ʹ), miR-107: (Forward, 5ʹ-AGCAGCATTGTACAGGG-3ʹ and Reverse, 5ʹ-GAATACCTCGGACCCTGC-3ʹ), Rab10: (Forward, 5ʹ-CACCGGATCGGGGATTCCGGAGTGG-3ʹ and Reverse, 5ʹ-AAACCCACTCCGGAATCCCCGATCC-3ʹ), U6: (Forward, 5ʹ-GCUUCGGCAGCACAUAUACUAAAAU-3ʹ and Reverse, 5ʹ-CGCUUCACGAAUUUGCGUGUCAU-3ʹ) and GAPDH: (Forward, 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ and Reverse, 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ).
Cell Proliferation Assay
Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) was used to detect the cell viability of the transfected TE-1 and KYSE30 cells according to its instructions. Briefly, TE-1 and KYSE30 cells (2×104 per well) were added in 96-well plate and cultivated for 24 hrs. Following transfection and 0-hr, 24-hr, 48-hr, or 72-hr incubation, CCK8 solution was injected into each well and incubated for another 3 hrs. The colorimetric analysis at 450 nm was measured using a spectrophotometer (Multiskan MK3; Thermo Fisher Scientific).
Transwell Assays
Transwell assay was used to detect the migration and invasion abilities in vitro. 24-well Transwell chambers (Corning, Tewksbury, MA, USA) with polycarbonate membrane were chosen in this assay. For migration, the lower chamber added with RPMI-1640 medium containing 10% FBS, and the upper chamber supplemented with transfected cells in serum-free medium. After 48-hr incubation, cells on the lower side of the membrane were fixed with 4% methanol and stained with 0.1% crystal violet. The cells in 10 random selected fields were counted using a light microscope (Olympus, Japan). For invasion, the protocol was similar to migration, while the upper chamber coated with a matrigel matrix (BD Biosciences, San Jose, CA, USA).
Dual-Luciferase Reporter Assay
The interactions between miR-107 and LINC00152 or Rab10 were predicted by starBase v2.0 (http://starbase.sysu.edu.cn) and TargetScan (http://www.targetscan.org), respectively. The wild type and mutant sequences of LINC00152 and Rab10 3ʹ-untranslated regions (3ʹUTR) were cloned and then inserted into luciferase vector psiCHECK2 (Promega, Madison, WI, USA), named as LINC00152 WT, LINC00152 MUT, Rab10 3ʹUTR- WT or Rab10 3ʹUTR-MUT. Subsequently, the luciferase reporter and miR-107 or miR-NC were co-transfected into TE-1 and KYSE30 cells. The luciferase reporter activity was measured using dual-luciferase reporter assay kit (Promega).
RNA Immunoprecipitation (RIP) Assay
For RIP assay, EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) were utilized to verify the interaction between miR-107 and LINV00152 according to its instructions. In brief, the TE-1 and KYSE30 cells lysate samples were incubated with RIP buffer containing magnetic beads linked with anti-Ago2 or anti-IgG. After elution by protease K, the level of LINC00152 was measured by qRT-PCR.
Western Blot Assay
The protein in TE-1 and KYSE30 cells was extracted using RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific) and the concentration was assessed by BCA Protein Assay Kit (Beyotime, Shanghai, China). Subsequently, the protein samples were separated by sodium dodecyl sulfonate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene fluoride (PVDF) membrane (GE Healthcare, Piscataway, NJ, USA). The membrane was blocked in no-fat milk for 4 hrs and then incubated with primary antibody overnight at 4°C. The membrane was incubated with secondary antibody at 37°C for another 2 hrs. RapidStep ECL Reagent (Millipore) was used to detect the fluorescence intensity. Rab10 (1:1000, ab230261), β-actin (1:1000, ab8227) and secondary antibody (1:25,000; ab97051) antibodies were obtained from Abcam (Cambridge, MA, USA).
Mice Model Experiments
The animal experiments were approved by the Animal Care Committee of the Third Hospital of Hebei Medical University in accordance with manual. Lentiviral vectors (sh-LINC00152 and sh-NC) were purchased from Genecham. Then, the TE-1 cells (4×106) transfected with sh-LINC00152 or si-NC were injected into the nude mice obtained from Shanghai Laboratory Animal Company (SLAC; Shanghai, China). The tumor volume was measured every 7 days continued 28 days and calculated according to the formula:volume (mm3) = width2 × length/2. Following the tumor excision, the xenograft tumors were weighted and frozen for further study.
Statistical Analysis
All data were performed using GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA), and exhibited as mean ± standard deviation (SD) from three independent repeats. Differences in two groups were assessed by Student's t-test, while among multigroup were processed through one-way analysis of variance (ANOVA) followed by Tukey’s post hoc. P<0.05 was considered statistically significant.
Results
LINC00152 Was Upregulated In ESCC Tissues And Cell Lines
Plenty of evidence indicated that LINC00152 was an oncogene in various malignancies. To observe the role of LINC00152 in ESCC, qRT-PCR was performed to detect the level of LINC00152 in ESCC tissues. The results showed that the level of LINC00152 was evidently increased in ESCC tissues in contrast to that in adjacent normal tissues (Figure 1A). The Kaplan-Meier curves exhibited that the survival rate of ESCC patients with high level of LINC00152 was apparently lower than that in low LINC00152 group (Figure 1B). In addition, the level of LINC00152 in cell lines also presented that LINC00152 was highly expressed in ESCC cell lines (TE-1, KYSE30) compared to that in human esophageal epithelial cell lines (Het-1A) (Figure 1C). Taken together, these results revealed the level of LINC00152 was significantly elevated in ESCC.
The Depletion Of LINC00152 Suppressed Cell Proliferation, Migration, And Invasion In TE-1 And KYSE30 Cells
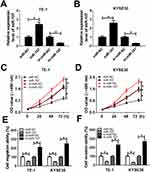
To observe the biological functions of LINC00152 in ESCC, loss-of-function assays were conducted in TE-1 and KYSE30 cells transfected with siRNAs (si-LINC00152#1, si-LINC00152#2 and si-LINC00152#3). The results displayed that LINC00152 was dramatically down-regulated in si-LINC00152-transfected TE-1 and KYSE30 cells, suggesting the high knockdown efficiency (Figure 2A and B). Subsequently, the CCK8 results indicated that the cell viability was strikingly decreased in TE-1 and KYSE30 cells transfected with si-LINC00152 related to that in si-NC or control group (Figure 2C and D). Also, the Transwell assay demonstrated that the abilities of cell migration and invasion of TE-1 and KYSE30 cells were remarkably reduced in si-LINC00152 group than that in si-NC or control group (Figure 2E and F). These data unraveled that LINC00152 knockdown blocked cell proliferation, migration and invasion in ESCC cells.
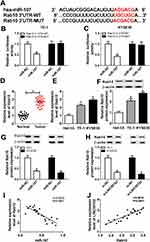
LINC00152 Directly Interacted With miR-107
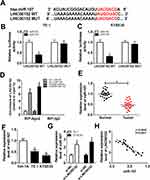
To investigate the mechanism of LINC00152 in ESCC, starBase v2.0 online database was used to search the putative target of LINC00152. The results showed that miR-107 had complementary sequences with LINC00152 (Figure 3A). Subsequently, dual-luciferase reporter assay was performed to verify this prediction. The luciferase activity of LINC00152 WT reporter was notably decreased in TE-1 and KYSE30 cells transfected miR-107 mimic compared with that in miR-NC treatment, while the luciferase activity of LINC00152 MUT reporter had no conspicuous change in two groups (Figure 3B and C). Furthermore, the RIP assay exhibited that Ago2 antibody enriched much more LINC00152 in TE-1 and KYSE30 cells transfected miR-107 compared with that in the IgG antibody group (Figure 3D). In addition, the qRT-PCR results illustrated that miR-107 was conspicuously reduced in ESCC tissues (Figure 3E) and cells (Figure 3F). MiR-107 was distinctly elevated in TE-1 and KYSE30 cells transfected with si-LINC00152 (Figure 3G). Meanwhile, the level of miR-107 was negatively linear correlated with the level of LINC00152 (Figure 3H). To sum, miR-107 was negatively interacted with LINC00152.
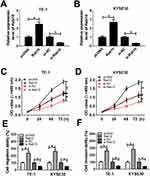
MiR-107 Retarded Cell Proliferation, Migration And Invasion In TE-1 And KYSE30 Cells
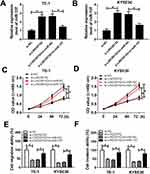
In order to explore the functions of miR-107 in ESCC, miR-107 mimics or in-miR-107 was transfected into TE-1 and KYSE30 cells. The transfection efficiency was confirmed, indicated by the striking enhancement of miR-107 in TE-1 and KYSE30 cells transfected with miR-107, or the apparent decrease of miR-107 in cells transfected with in-miR-107 (Figure 4A and B). The transfection of miR-107 contributed to the distinct reduction of cell viability TE-1 and KYSE30 cells related to that in miR-NC group, while the introduction of in-miR-107 showed the opposite trend (Figure 4C and D). Also, the migration and invasion abilities were showed the same trend. Briefly, the migration and invasion abilities were obviously declined in TE-1 and KYSE30 cells transfected with miR-107, while conspicuously elevated in in-miR-107-transfected TE-1 and KYSE30 cells (Figure 4E and F). These data illustrated that miR-107 retarded ESCC progression.
LINC00152 Promoted Cell Proliferation, Migration And Invasion By Targeting miR-107
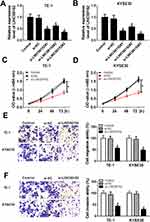
Based on the above results, we found that miR-107 was negatively interacted with LINC00152 in ESCC cells. To further implicate the interaction between LINC00152 and miR-107, we conducted the restoration experiment. The results showed that miR-107 was elevated in TE-1 and KYSE30 cells induced by si-LINC00152, while the promoted effect was mitigated by the emergence of miR-107 inhibitor (Figure 5A and B). Similarly, the transfection of in-miR-107 receded the constraint impact on cell viability in TE-1 and KYSE30 cells caused by si-LINC00152 (Figure 5C and D). Also, the migration and invasion abilities were suppressed by the LINC00152 knockdown in TE-1 and KYSE30 cells, but the introduction of miR-107 relieved the inhibitory effect (Figure 5E and F). These data suggested that LINC00152 accelerated cell proliferation, migration and invasion by targeting miR-107.
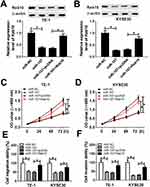
Rab10 Was A Target Of miR-107
In order to further understand the underlying biological role of miR-107 in ESCC, the putative target of miR-107 was searched by the TargetScan. The results presented that Rab10 3ʹUTR had complementary binding sites with miR-107 (Figure 6A). Following dual-luciferase reporter assay exhibited that the transfection of miR-107 mimics led to the notable downregulation of luciferase activity of Rab10 3ʹUTR-WT reporter, but had no influence on luciferase activity of Rab10 3ʹUTR-MUT reporter (Figure 6B and C).
Furthermore, the mRNA and protein levels of Rab10 were distinctly elevated in ESCC tissues and cells (Figure 6D–F). The protein level of Rab10 was significantly down-regulated in TE-1 and KYSE30 cells transfected with miR-107 mimics or si-LINC00152 (Figure 6G and H). In addition, the qRT-PCR results revealed that Rab10 was negatively linear correlated with miR-107, while LINC00152 and Rab10 presented the opposite trend (Figure 6I and J). These data unraveled that Rab10 was a target of miR-107, and its level was upregulated in ESCC.
Rab10 Accelerated Cell Proliferation, Migration And Invasion In TE-1 And KYSE30 Cells
To explore the functions of Rab10 in ESCC, Rab10 or si-Rab10 was transfected into TE-1 and KYSE30 cells. The qRT-PCR results showed that the level of Rab10 was markedly increased in TE-1 and KYSE30 cells transfected with Rab10, or dramatically decreased in cells transfected with si-Rab10, indicating the transfection efficiency (Figure 7A and B). The transfection of si-Rab10 resulted in the significant decline of cell viability TE-1 and KYSE30 cells, while Rab10 showed the opposite trend (Figure 7C and D). Also, the migration and invasion abilities were showed the same trend with that of cell viability. Briefly, the migration and invasion abilities were evidently upregulated in TE-1 and KYSE30 cells transfected with Rab10, while strikingly reduced in si-Rab10-transfected TE-1 and KYSE30 cells (Figure 7E and F). These data illustrated that Rab10 facilitated ESCC progression.
miR-107 Inhibited Cell Proliferation, Migration And Invasion In ESCC Cells By Targeting Rab10
Next, the restoration experiment was performed to illuminate the functions between miR-107 and Rab10. The transfection of miR-107 mimics resulted in the striking reduction of the protein level of Rab10 in TE-1 and KYSE30 cells, but Rab10 overexpression mitigated the inhibitory effect (Figure 8A and B). Similarly, the transfection with Rab10 regained cell viability in TE-1 and KYSE30 cells inhibited by miR-107 mimics (Figure 8C and D). The Transwell assay presented that Rab10 overexpression alleviated the suppressive effects on migration and invasion abilities in TE-1 and KYSE30 cells induced by miR-107 (Figure 8E and F). Taken together, miR-107 blocked cell proliferation, migration and invasion in ESCC cells by targeting Rab10.
LINC00152 Positively Regulated Rab10 Expression By Sponging miR-107
To further manifested the regulatory network in LINC00152, miR-107 and Rab10, the TE-1 and KYSE30 cells were co-transfected with si-LINC00152 and in-miR-107. The results showed that the mRNA and protein levels of Rab10 were markedly decreased in si-LINC00152-transfected TE-1 and KYSE30 cells, and the introduction of miR-107 mimics partly recovered the level of Rab10 (Figure 9A and B). These data implicated that LINC00152 regulated the expression of Rab10 mediated by miR-107.
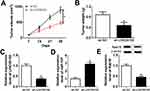
LINC00152 Knockdown Impeded Xenograft Growth In Vivo
To confirm the functional effects of LINC00152 in vivo, mice model was established. TE-1 cells transfected with sh-LINC00152 or sh-NC were injected into nude mice. As shown in Figure 10A and B, the tumor volume and weight were both significantly reduced in sh-LINC00152 group related to that in sh-NC group. Furthermore, the level of LINC00152 was drastically down-regulated and miR-107 was evidently elevated in sh-LINC00152 group compared with that in sh-NC group (Figure 10C and D). Also, the protein level of Rab10 was distinctly decreased in sh-LINC00152 group (Figure 10E). These results illustrated that the depletion of LINC00152 hampered xenograft growth in vivo.
Discussion
Esophageal squamous cell carcinoma (ESCC) is a common malignant cancer in East Asia. Emerging evidence has manifested that lncRNAs played a crucial role in many types of cancers.13–15 In this study, we aimed to find the regulatory network of LINC00152 in ESCC. Our data showed that LINC00152 regulated Rab10 to impel cell proliferation, migration and invasion in ESCC by sponging miR-107.
Recent study indicated that the dysregulation of LINC00152 was found in various malignancies. For instance, a study in oral carcinoma showed that the level of LINC00152 was distinctly upregulated in oral carcinoma tissues.16 Another reporter manifested that LINC00152 was remarkably elevated in ovarian cancer tissues and cell lines, and its silencing inhibited cell proliferation while induced cell apoptosis in ovarian cancer.17 In the present study, we verified that LINC00152 was highly expressed in ESCC tissues and cells. Furthermore, LINC00152 knockdown suppressed cell proliferation, migration and invasion in ESCC. In addition, LINC00152 blocked xenograft tumor growth in vivo. These results confirmed that LINC00152 played a vital role in ESCC.
LncRNAs have been documented to function as competing endogenous RNAs (ceRNAs) to sponge miRNAs, thus resulting in the mRNAs abnormal expression.18 The aberrant expression of miR-107 has been reported in various cancers including ESCC. In this study, we found that the level of miR-107 was obviously reduced in ESCC tissues and cells (TE-1 and KYSE30). LINC00152 was validated as a sponge for miR-107. The inhibitor of miR-107 promoted cell proliferation and metastasis. Moreover, miR-107 overexpression alleviated the inhibitory effects on cell proliferation, migration and invasion induced by the depletion of si-LINC00152. In agreement with our data, a study in cervical cancer that miR-107 was down-regulated and retarded cell growth mediated by lncRNA DLG1-AS1.19 The similar results of miR-107 were reported in pancreatic cancer.20 Our results of miR-107 in ESCC were consistent with previous reports.10,21 These data demonstrated that LINC00152 promoted cell proliferation, migration and invasion in ESCC by sponging miR-107.
Rab10 has been known as a member in RAB family of small GTPases, and its aberrant expression was associated with malignancy.11 For example, a study in osteosarcoma showed that Rab10 knockdown inhibited cell proliferation and migration.22 In ESCC, a report indicated that Rab10 depletion blocked cell proliferation, migration and invasion.12 In this study, the qRT-PCR results showed that Rab10 was notably increased in ESCC tissues and cells. Rab10 was negatively interacted with miR-107. The silencing of Rab10 impeded cell growth and metastasis of ESCC cells. Moreover, Rab10 overexpression mitigated the inhibitory effects on cell proliferation, migration and invasion induced by miR-107. Their data illustrated that miR-107 suppressed cell proliferation, migration and invasion by targeting Rab10 in ESCC.
In conclusion, all the results unraveled that LINC00152/miR-107/Rab10 regulated cell proliferation, migration and invasion in ESCC. Moreover, it has been reported that LINC00152 regulated cell proliferation, migration and apoptosis via miR-153-3p/FYN axis in ESCC.8 Combined with the above regulatory network, the role of LINC00152 may play crucial role in ESCC and LINC00152/miR-107/Rab10 new regulatory pathway provides new perspective into ESCC progression, thus may provide novel therapeutic target for ESCC patients.
Ethics Approval And Consent To Participate
The present study was approved by the ethical review committee of the Third Hospital of Hebei Medical University. Written informed consent was obtained from all enrolled patients.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64(3):381–387. doi:10.1136/gutjnl-2014-308124
2. Zeng H, Zheng R, Guo Y, et al. Cancer survival in China, 2003-2005: a population-based study. Int J Cancer. 2015;136(8):1921–1930. doi:10.1002/ijc.29227
3. Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349(23):2241–2252. doi:10.1056/NEJMra035010
4. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi:10.1038/nrg3074
5. Neumann O, Kesselmeier M, Geffers R, et al. Methylome analysis and integrative profiling of human HCCs identify novel protumorigenic factors. Hepatology (Baltimore, Md). 2012;56(5):1817–1827. doi:10.1002/hep.25870
6. Yu Y, Yang J, Li Q, Xu B, Lian Y, Miao L. LINC00152: a pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017;50(4). doi:10.1111/cpr.12349
7. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14(1):42–54. doi:10.1016/j.gpb.2015.09.006
8. Liu D, Gao M, Wu K, Zhu D, Yang Y, Zhao S. LINC00152 facilitates tumorigenesis in esophageal squamous cell carcinoma via miR-153-3p/FYN axis. Biomed Pharmacother. 2019;112:108654. doi:10.1016/j.biopha.2019.108654
9. Zhao X, Li H, Wang L. MicroRNA-107 regulates autophagy and apoptosis of osteoarthritis chondrocytes by targeting TRAF3. Int Immunopharmacol. 2019;71:181–187. doi:10.1016/j.intimp.2019.03.005
10. Sharma P, Saini N, Sharma R. miR-107 functions as a tumor suppressor in human esophageal squamous cell carcinoma and targets Cdc42. Oncol Rep. 2017;37(5):3116–3127. doi:10.3892/or.2017.5546
11. Chua CEL, Tang BL. Rab 10-a traffic controller in multiple cellular pathways and locations. J Cell Physiol. 2018;233(9):6483–6494. doi:10.1002/jcp.26503
12. Ding N, Sun X, Wang T, Huang L, Wen J, Zhou Y. miR‑378a‑3p exerts tumor suppressive function on the tumorigenesis of esophageal squamous cell carcinoma by targeting Rab10. Int J Mol Med. 2018;42(1):381–391.doi:10.3892/ijmm.2018.3639
13. Huynh NP, Anderson BA, Guilak F, McAlinden A. Emerging roles for long noncoding RNAs in skeletal biology and disease. Connect Tissue Res. 2017;58(1):116–141. doi:10.1080/03008207.2016.1194406
14. Li H, Ma SQ, Huang J, Chen XP, Zhou HH. Roles of long noncoding RNAs in colorectal cancer metastasis. Oncotarget. 2017;8(24):39859–39876. doi:10.18632/oncotarget.16339
15. Wang J, Sun J, Wang J, et al. Long noncoding RNAs in gastric cancer: functions and clinical applications. Onco Targets Ther. 2016;9:681–697. doi:10.2147/OTT.S95412
16. Chen M, Xu X, Ma H. Identification of oncogenic long noncoding RNAs CASC9 and LINC00152 in oral carcinoma through genome-wide comprehensive analysis. Anticancer Drugs. 2019;30(4):356–362. doi:10.1097/CAD.0000000000000725
17. Chen P, Fang X, Xia B, Zhao Y, Li Q, Wu X. Long noncoding RNA LINC00152 promotes cell proliferation through competitively binding endogenous miR-125b with MCL-1 by regulating mitochondrial apoptosis pathways in ovarian cancer. Cancer Med. 2018;7(9):4530–4541. doi:10.1002/cam4.1547
18. Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi:10.1016/j.semcdb.2014.05.015
19. Rui X, Xu Y, Huang Y, Ji L, Jiang X. lncRNA DLG1-AS1 promotes cell proliferation by competitively binding with miR-107 and up-regulating ZHX1 expression in cervical cancer. Cell Physiol Biochem. 2018;49(5):1792–1803. doi:10.1159/000493625
20. Imamura T, Komatsu S, Ichikawa D, et al. Depleted tumor suppressor miR-107 in plasma relates to tumor progression and is a novel therapeutic target in pancreatic cancer. Sci Rep. 2017;7(1):5708. doi:10.1038/s41598-017-06137-8
21. Sharma P, Saraya A, Gupta P, Sharma R. Decreased levels of circulating and tissue miR-107 in human esophageal cancer. Biomarkers. 2013;18(4):322–330. doi:10.3109/1354750X.2013.781677
22. Jiang W, Liu J, Xu T, Yu X. MiR-329 suppresses osteosarcoma development by downregulating Rab10. FEBS Lett. 2016;590(17):2973–2981. doi:10.1002/1873-3468.12337
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.